Abstract
Background
Studying comorbidities of disorders is important for detection and prevention. For discovering frequent patterns of diseases we can use retrospective analysis of population data, by filtering events with common properties and similar significance. Most frequent pattern mining methods do not consider contextual information about extracted patterns. Further data mining developments might enable more efficient applications in specific tasks like comorbidities identification.
Methods
We propose a cascade data mining approach for frequent pattern mining enriched with context information, including a new algorithm MIxCO for maximal frequent patterns mining. Text mining tools extract entities from free text and deliver additional context attributes beyond the structured information about the patients.
Results
The proposed approach was tested using pseudonymised reimbursement requests (outpatient records) submitted to the Bulgarian National Health Insurance Fund in 2010–2016 for more than 5 million citizens yearly. Experiments were run on 3 data collections. Some known comorbidities of Schizophrenia, Hyperprolactinemia and Diabetes Mellitus Type 2 are confirmed; novel hypotheses about stable comorbidities are generated. The evaluation shows that MIxCO is efficient for big dense datasets.
Conclusion
Explicating maximal frequent itemsets enables to build hypotheses concerning the relationships between the exogeneous and endogeneous factors triggering the formation of these sets. MixCO will help to identify risk groups of patients with a predisposition to develop socially-significant disorders like diabetes. This will turn static archives like the Diabetes Register in Bulgaria to a powerful alerting and predictive framework.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Motivation
Studying comorbidities of disorders is important for detection and prevention. For discovering frequent patterns of diseases we can use retrospective analysis of population data, by filtering events with common properties and similar significance. Two major approaches to pattern search are: (i) frequent pattern mining (FPM) viewing the events (objects) as unordered sets and (ii) frequent sequence mining (FSM). Most FPM and FSM methods do not consider contextual information about extracted patterns; usually they build a prefix tree, which is huge and difficult to manipulate in memory when big data is processed [1]. Further developments of data mining (DM) might enable more efficient applications in specific tasks e.g. identification of relations between different unrelated diseases—so called comorbidity.
Clinical narratives are an underused data source that has much greater research potential than is currently realized. Biomedical scientists increasingly use text mining to extract important entities from medical texts and integrate them in various DM studies. However automated analysis of clinical texts is most successful for English while only fragmented components exist for the other languages. So advancing the biomedical Natural Language Processing (NLP) for under-resourced languages is another challenge to be met in order to improve DM achievements in knowledge discovery.
We propose a cascade data mining approach for frequent pattern mining enriched with context information, including a new algorithm MIxCO for maximal frequent patterns mining. Text mining tools extract entities from free text and deliver additional context attributes beyond the structured information about the patients. NLP for Bulgarian delivers entities from Outpatient Records (ORs) free texts. Novel hypotheses are generated to discover stable comorbidities and to confirm known ones. The experiments explicate some population specific comorbidities. We also discuss the effects of age, gender and demographics on these comorbidities.
The paper is structured as follows. Section 2 presents related work, Sect. 3—the data we use, Sect. 4—the methods. Section 5 discusses current experiments and their medical interpretation. Section 6 sketches further work and the conclusion.
Related work
The concept of frequent itemsets is introduced by Agrawal et al. [2]. Methods for solving FPM vary from the naive BruteForce and Apriori algorithms, where the search space is organized as a prefix tree, to Eclat/dEclat algorithm that uses tidsets directly for support computation, by processing prefix equivalence classes [1]. Another efficient algorithm is FPGrowth (Frequent Pattern Tree Approach). Using the generated frequent patterns we can later generate association rules. Most FPM algorithms generate all possible frequent patterns (FPs). The search space grows exponentially with the number of items. Summarized information for data relations can be extracted as maximal frequent itemsets (MFI). The condensed information not only accelerates the process, reducing redundancy, but also decreases significantly the number of FPs for post-analysis. All classic algorithms for FPM can be modified for MFI search, by checking for maximality at each step. There are some especially designed algorithms for MFI search, e.g. the MFCS algorithm which combines top-down and bottom-up [3]. The GenMax algorithm that uses a vertical database, diffsets and optimizations by checking whether the union of all itemsets is included already in some maximal itemset and then pruning the branch [4]. The FPMax algorithm is based on FP-trees by extending FP-growth algorithm [5]. MAFIA uses depth-first traversal of the itemset lattice with effective pruning mechanisms which is quite good especially when the database itemsets are very long [6].
NLP for English clinical texts made significant progress in algorithm development and resource construction since 2000. Open-source tools like cTAKESFootnote 1 extracts information from clinical free text. Another open source system is HITEx (Health Information Text Extraction) which extracts variables of interest from narratives [7]. Despite the limitations, the NLP importance as a supporting technology will grow due to its constant improvements [8].
Studies on multimorbidity are a great challenge given the mismatch between the high prevalence of this condition and relatively smaller number of research papers [9], which is partly due to lack of data. Machine learning (ML) is the basic technology used in such studies. For instance, four ML techniques (logistic regression, k-nearest neighbors, multifactor dimensionality reduction and support vector machines) were applied to assess risks for diabetes, hypertension and their comorbidity in a cohort of 270,172 hospital visitors (89,858 diabetic, 58,745 hypertensive and 30,522 comorbid patients) in Kuwait, with accuracy > 85% (for diabetes) and > 90% (for hypertension) [10]. An original approach for predicting a comorbid medical condition incidence and progression of medical conditions, using self-posted data available on patient-oriented social media sites, is presented in [11]. The similarity between patient postings is calculated and the risk of a condition is derived thus producing a ranked list of medical conditions for each patient. An algorithm to build medical condition progression trajectories is suggested. The condition incidence model predicts future conditions with coverage of 48% (top-20) and 75% (top-100).
Materials
The data repository we use currently contains more than 262 million pseudonymised ORs submitted to the Bulgarian National Health Insurance Fund (NHIF) in 2010-2016 for more than 5 mln citizens yearly. In Bulgaria ORs are produced by the General Practitioners and the Specialists from Ambulatory Care for every contact with the patient. Despite their primary accounting purpose the ORs summarise sufficiently the case and motivate the requested reimbursement. ORs are semi-structured files with predefined XML-format. The structured XML fields provide useful information: date and time of the visit; pseudonymised personal data, age, gender; pseudonymised visit-related information; diagnoses in ICD-10Footnote 2; NHIF drug codes for medications that are reimbursed; a code if the patient needs special monitoring; a code concerning the need for hospitalization; several codes for planned consultations, lab tests and medical imaging. The free text OR fields, listed in Table 1, are processed by our NLP tools.
Methods
The system architecture is shown on Fig. 1. Text mining modules convert the raw text to structured data. We developed a drug extractor using regular expressions to describe linguistic patterns [12], it handles 2239 drug names included in the NHIF nomenclatures. For extraction of clinical test data (body mass index—BMI, weight, blood pressure etc.) we designed a Numerical value extractor [13].
We search for as many as possible associations between chronic diseases.Footnote 3 A tabular method using a vertical database is proposed, with depth-first traversal as well as set intersection and diffsets. Further processing of the MFI is applied to remove diagnostic related groups. Some context information is added to each MFI to study comorbidities. This information is presented as attribute-value tuples for each patient; the post-processing identifies the importance of different attributes for each MFI.
Mining maximal frequent itemsets
For the collection S of ORs we extract the set of all different patient identifiers \(P = \left\{ {p_{1} ,p_{2} , \ldots ,p_{N} } \right\}\). This set corresponds to transaction identifiers (tids) and we call them pids (patient identifiers). We consider each patient visit to a doctor as a single event. For each patient \(p_{i} \in P\) an event sequence of tuples \(\langle event,timestamp \rangle\) is generated: \(E\left( {p_{i} } \right) = \left(\langle {e_{1} ,t_{1}\rangle ,\langle e_{2} ,t_{2} \rangle, \ldots ,\langle e_{{k_{i} }} ,t_{{k_{i} }} } \rangle \right), i = \overline{1,N}\). Let \({\mathcal{E}}\) be the set of all possible events and \({\mathcal{T}}\) be the set of all possible timestamps. Let \({\mathcal{C}} = \left\{ {c_{1} ,c_{2} , \ldots ,c_{p} } \right\}\) be the set of all chronic diseases, which we call items. Each subset of \(X \subseteq {\mathcal{C}}\) is called an itemset. We define a projection function \(\pi :\left( {{\mathcal{E}} \times {\mathcal{T}}} \right)^{N} \to {\mathcal{C}}^{N}\): \(\pi \left( {E\left( {p_{i} } \right)} \right) = C\left( {p_{i} } \right) = \left( {c_{{1{\text{i}}}} ,c_{{2{\text{i}}}} , \ldots ,c_{{\text{mi} }} } \right)\), such that for each patient \(p_{i} \in P\) the projected time sequence contains only the first occurrence (onset) of each chronic disorder recorded in \(E\left( {p_{i} } \right)\). Let \(D \subseteq P \times 2^{{\mathcal{C}}}\) be the set of all itemsets in our collection after projection \(\pi\) in the format \(\langle pid,itemset \rangle\). We will call \(D\) database. We are looking for itemsets \(X \subseteq {\mathcal{C}}\) with frequency (\({ \sup }(X)\)) above given \(minsup.\) Let \({\mathcal{F}}\) denote the set of all frequent itemsets, i.e. \({\mathcal{F}} = {\text{\{ }}X | X \subseteq {\mathcal{C} }\;and\;{ \sup }(X) \ge minsup\}\). A frequent itemset \(X \in {\mathcal{F}}\) is called maximal if it has no frequent supersets. Let \({\mathcal{M}}\) denote the set of all maximal frequent itemsets, i.e. \({\mathcal{M}} = {\text{\{ }}X | X \in {\mathcal{F}}\;\text{ }and\; {\nexists } Y \in {\mathcal{F}},\text{ }\;such \;that \;X \subset Y\}\). Let \(2^{X}\) denote the power set (set of all subsets) of itemset \(X.\) Then each subset of \(X \in {\mathcal{F}}\) is also frequent itemset, i.e. \(\forall Y \in 2^{X} \; implies \;that \;Y \in {\mathcal{F} }\). For each item \(c \in {\mathcal{C}}\) we define the set called pidset: \(p\left( {\text{c}} \right) = {\text{\{ }}p_{i} | \langle p_{i} ,C\left( {p_{i} } \right)\rangle \in D \;and\; {\text{c}} \in C\left( {p_{i} } \right)\}\).
We preprocess the database \(D\) by generating pidsets and transform it to vertical database \(D^{V}\): \(D^{V} = { \langle \text{\{ c}},p\left( c \right) \rangle |c \in {\mathcal{C} }\}\). Let \(w \in {\mathcal{C}}\), we define projection \(P_{w}\) of the database \(D^{V}\) by pidsets intersection: \(P_{w} \left( {D^{V} } \right) = {\text{\{ }} \langle {\text{c}},p^{\prime}\left( c \right)\rangle | \langle{\text{c}},p\left( c \right) \rangle \in D^{V} , {\text{c}} \ne {\text{w }}\;\;and\; p^{\prime}\left( c \right) = p\left( c \right)\mathop \cap \nolimits {\text{p}}({\text{w}})\}\) and its complement by pidsets difference: \(\overline{{P_{w} \left( {D^{V} } \right)}} = {\text{\{}} \langle {\text{c}},p^{\prime\prime}\left( c \right) \rangle|\langle {\text{c}},p\left( c \right) \rangle\in D^{V} , {\text{c}} \ne {\text{w }}\;and\; p^{\prime\prime}\left( c \right) = p\left( c \right) - {\text{p}}({\text{w}})\}\). Let \(f(c)\) denotes the frequency of item \(c \in {\mathcal{C}}\) in database \(D^{V}\). An item \(w \in {\mathcal{C}}\) is called weak, if there has no item in \({\mathcal{C}}\) with support lower than \(f(w)\), i.e. \({\nexists } c \in {\mathcal{C}}\) such that \(f\left( c \right) < f(w)\).
Algorithm MIxCO (MIning COmorbidity)
Assume that the set of all maximal frequent itemsets \({\mathcal{M}}\) is initially the empty set. We reduce the database \(D^{V}\) by deleting all tuples that contain items with support below \(minsup\) and process further the obtained database \(D^{{V^{\prime}}}\). Obviously the maximal frequent itemsets will contain as many as possible items, thus they must contain also items with low frequency. In order to identify maximal frequent itemsets we start from the weakest item \(w \in {\mathcal{C}}\) in \(D^{{V^{\prime}}}\). There are two cases: either a maximal frequent itemset \(X\) contains \(w\), or it does not contain it. Thus we need to split \(D^{{V^{\prime}}}\) in two subsets by projections \(P_{w} \left( {D^{{V^{\prime}}} } \right)\) and \(\overline{{P_{w} \left( {D^{{V^{\prime}}} } \right)}}\). We apply recursively the algorithm MIxCO for searching all maximal frequent itemsets in \(P_{w} \left( {D^{{V^{\prime}}} } \right)\). Let the result set of all maximal frequent itemsets in \(P_{w} \left( {D^{{V^{\prime}}} } \right)\) be \({\mathcal{M}}_{w}\). We add to each of them the item \(w\): \({\mathcal{M}}_{w}^{'} = {\text{\{ }}Y | X \in {\mathcal{M}}_{w} , Y = X\mathop \cup \nolimits \left\{ w \right\}\}\) and obtain the maximal frequent itemsets that contain \(w\). Let \({\mathcal{B}}\) be the set of all members of \(P_{w} \left( {D^{{V^{\prime}}} } \right)\) that were reduced from the algorithm MIxCO due to low frequency (bellow the \(minsup\)). These items cannot be reduced from further considerations because they have low frequency in combination with \(w\), but support above \(minsup\) in the entire database \(D^{{V^{\prime}}}\) and they can be members of maximal frequent itemsets that do not contain \(w\). We update \(\overline{{P_{w} \left( {D^{{V^{\prime}}} } \right)}}\) by adding those itemsets that contain members of \({\mathcal{B}}\):
\(\overline{{P_{w} \left( {D^{{V^{\prime}}} } \right)}}^{U} = \overline{{P_{w} \left( {D^{{V^{\prime}}} } \right)}} \mathop \cup \nolimits {\text{\{ }}\langle c,y\mathop \cup \nolimits \left( {z\mathop \cap \nolimits p\left( x \right)} \right)\rangle |\langle x,p\left( x \right) \rangle \in {\mathcal{B}}, \langle c,z\rangle \in P_{w} \left( {D^{{V^{\prime}}} } \right),\;\langle c,y\rangle \in \overline{{P_{w} \left( {D^{{V^{\prime}}} } \right)}} \} .\) We apply recursively the algorithm MIxCO for searching all maximal frequent itemsets in \(\overline{{P_{w} \left( {D^{{V^{\prime}}} } \right)}}^{U}\). Let the result set of all maximal frequent itemsets in \(\overline{{P_{w} \left( {D^{{V^{\prime}}} } \right)}}^{U}\) be \(\overline{{{\mathcal{M}}_{w} }}\). Then the result set of all maximal frequent itemsets of the database \(D\) is the union \({\mathcal{M}} = {\mathcal{M}^{\prime}}_{w} \mathop \cup \nolimits \overline{{{\mathcal{M}}_{w} }} .\) Finally we reduce \({\mathcal{M}}\) by removing all frequent patterns that are not maximal, if any.
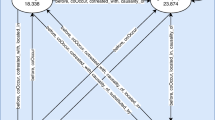
We illustrate \(MIxCO\) by a synthetic example (Fig. 2). Itemsets of ICD-10 codes for 10 patients are presented. For each ICD-10 code (F20, E11, I11, M17, I20, E66, J44) is generated a set of pids, i.e. \(DV\). We apply reduction for \(minsup = 3\) and obtain \(B = \{ \langle M17,\{ 2,4\}\rangle , \langle E66,\{ 10\} \rangle, \langle J44,\{ 5\} \rangle\}\). The weakest item of the new set \(DV^{\prime}\) is \(w = I20\). On the next step we partition \(DV^{\prime}\) into two subsets by projection \(P_{I20} \left( {DV^{\prime}} \right)\) and \(\overline{{P_{I20} \left( {DV^{\prime}} \right)}}\). First we start processing \({P_{I20} \left( {DV^{\prime}} \right)}\) and apply reduction with \(B^{\prime} = \{\langle {\text{I}}11,\{ 8\} \rangle\}\). The weakest item in the resuced set \(P_{I20} \left( {DV^{\prime\prime}} \right)\) is \(w = F20\). We apply projection and obtain to subsets \(P_{F20} \left( {P_{I20} \left( {DV^{\prime\prime}} \right)} \right)\) and \(\overline{{P_{F20} \left( {P_{I20} \left( {DV^{\prime\prime}} \right)} \right)}}\). Because for \(P_{F20} \left( {P_{I20} \left( {DV^{\prime\prime}} \right)} \right)\) no reduction is applied and its cardinality is 1, we return the frequent itemset \({\mathcal{M}} = \{ \{ F20, E11, I20\} \}\), which contains items from both projections F20 and I20 and the only left item E11 in the later subset. The subset \(\overline{{P_{F20} \left( {P_{I20} \left( {DV^{\prime\prime}} \right)} \right)}}\) is empty and the algorithm terminates processing the subset \(P_{I20} \left( {DV^{\prime}} \right)\). We continue by processing \(\overline{{P_{I20} \left( {DV^{\prime}} \right)}}\) and update it by the reduced data from \(B^{\prime}\). No further reductions are applied to the updated set \(\overline{{P_{I20} \left( {DV^{\prime\prime}} \right)}} ,\) because all subsets have support above minsup. The weakest item in \(\overline{{P_{I20} \left( {DV^{\prime\prime}} \right)}}\) is \(w = F20\). We apply projection and obtain to subsets \(P_{F20} \left( {\overline{{P_{I20} \left( {DV^{\prime\prime}} \right) }} } \right)\) and \(\overline{{P_{F20} \left( {\overline{{P_{I20} \left( {DV^{\prime\prime}} \right) }} } \right)}}\). For \(P_{F20} \left( {\overline{{P_{I20} \left( {DV^{\prime\prime}} \right) }} } \right)\) no reduction is applied and its weakest item is \(w = E11\). We apply projection and obtain to subsets \(P_{E11} \left( {P_{F20} \left( {\overline{{P_{I20} \left( {DV^{\prime\prime}} \right) }} } \right)} \right)\) and \(\overline{{P_{E11} \left( {P_{F20} \left( {\overline{{P_{I20} \left( {DV^{\prime\prime}} \right) }} } \right)} \right)}}\) and so on. The frequent itemsets \({\mathcal{M}} = \{ \left\{ {F20, E11, I20} \right\}, \left\{ {F20, E11} \right\}, \left\{ {F20, I11} \right\}, \{ E11, I11\} \}\), produced at the end, are presented in oval shapes in the leaves of the tree on Fig. 2. Finally we reduce the non maximal itemsets from \({\mathcal{M}}\), i.e. \(\left\{ {F20, E11} \right\} \subset \left\{ {F20, E11, I20} \right\}\), presented in shadow in Fig. 2. The result set \({\mathcal{M}} = \{ \left\{ {F20, E11, I20} \right\}, \left\{ {F20, I11} \right\},\{ E11, I11\} \}\) contain maximal frequent itemsets only.

Context information
Comorbidities need to be studied in the context where they occur so we add semantic attributes to each event—patient demographics, age and gender, treatment, status etc.
We define a set of attributes of interest \(A = \{ a_{1} ,a_{2} , \ldots ,a_{k} \}\). Context Q for some patient \(p_{i} \in P\) is defined as the set of attribute-value pairs from patient profile information: \(Q\left( {p_{i} } \right) = \{ \langle a_{1} ,q_{1}\rangle ,\langle a_{2} ,q_{2}\rangle , \ldots ,\langle a_{k} ,q_{k}\rangle \}\). In order to decrease the number of possible values of attributes we apply some aggregation of data. For instance age value is categorized according to the World Health Organization (WHO) standard age groups.Footnote 4 Data for body mass index (BMI) are also categorized according to the WHOFootnote 5 standard classification—underweight, normal weight, overweight, obesity. For some data concerning demographic information, like region ID we have large number of distinct values. For such data we add also some additional properties concerning background information for the region—e.g. whether it is south, north, west, east, central, northwest etc., and mountain, river, sea, thermal spring, urban region etc.
From \(Q\left( {p_{i} } \right)\) we generate a feature vector \(v\left( {p_{i} } \right) = \left( {v_{1i} ,v_{2i} , \ldots ,v_{mi} } \right)\), where each attribute \(a_{j} \in A\) with \(N_{j}\) possible values is represented by \(N_{j}\) consecutive positions in the vector. For the set of maximal frequent itemset \({\mathcal{M}}\) with cardinality \(\left| {\mathcal{M}} \right| = {\text{K}}\) we have \({\text{K}}\) classes of comorbidities. We apply classification of multiple classes in order to generate rules for each comorbidity class. We use SVM and optimization based on block minimization method described by Yu et al. [14].
Experiments and medical relevance
MIxCO algorithm evaluation
Some evaluation experiments were performed for MixCO and FPMax algorithms with two databases A and B. The number of transaction in both collections is 11,345, but A is very dense, and in contrast B is very sparse. The number of items in A is 4337, and in B is 3412. Table 2 shows the execution time in milliseconds for a relative minsup between 0.01 and 0.05.
The evaluation results show that FP-Max outperforms MIxCO for big sparse databases. In contrast MIxCO shows better results for big dense databases.
Comorbidity identification
The term “comorbid” here means “indicating two or more medical conditions existing simultaneously regardless of their causal relationship”. One comprehensive study of the possible relations between comorbid diseases is [15]. The authors describe 13 comorbid models, also known as “NK models”, which allow to examine the etiology of the comorbidity between disorders and to predict mortality and other outcomes.
Our experiments for pattern search are made on five OR collections that are used as training and test corpora (Table 3). They contain data about patients suffering from Schizophrenia (ICD-10 code F20), Hyperprolactinaemia (ICD-10 code E22.1), and Diabetes Mellitus Type 2 (ICD-10 code E11). Schizophrenia and Diabetes Mellitus are chronic diseases with a variety of complications that are also chronic diseases. The collections are extracted by using a Business Intelligence Tool (BITool) [13] from the repository of ORs for approx. 5 million patients for a 3-years period.
The minsup value was set as relative minsup function of the ration between the number of patients and ORs. It is approximately between 0.015% for S2 and S3 and 0.005% for S1. This is a rather small minsup value that will guarantee coverage even for more rare chronic diseases but with sufficient support.
The noise in the data is not taken into account. We do not discuss the correctness of the clinical data from medical point of view. The average number of ORs per patient is distributed almost evenly in the collections S1–S3: 12.2 (set S1), 9.85 (S2) and 14.5 (S3) and each patient has several visits each year. On the other hand the collections are almost complete and cover the population in Bulgaria for these period.
The experimental collections were carefully selected. The association between Schizophrenia, Hyperprolactinemia, and Diabetes Mellitus Type 2 is well known so it is easier to assess the novelty of discovered comorbidities corresponding to the extracted maximal frequency itemsets.
Comorbidity interpretation in psychiatric diseases has specific aspects because in mental health comorbidity does not necessarily imply the presence of multiple diseases. It usually is the result of imprecisely distinguished mental illnesses and inability to supply a single diagnosis that accounts for all symptoms. For example in collection S1 the support of itemset \(\{ F20, F31\}\) is 871, where \(F31\) is Bipolar affective disorder and \(F20\) is Schizophrenia. Despite this imperfection, we see that the longest maximal frequent itemsets overcome this problem. Table 4 contains diseases with ICD-10 codes I11 (Hypertensive heart disease with heart failure), I20 (Angina pectoris), I50 (Heart failure), I69 (Sequelae of cerebrovascular disease). The result is not quite surprising due to the well-studied comorbidity between Schizophrenia and Cardio-vascular diseases [15].
Interesting and unexpected results were found in the set of maximal frequent itemsets with size 5 (Table 5)—comorbidity with M17 Gonarthrosis (arthrosis of knee).
This correlation seems to be a new hypothesis: a search PubMed found only 3 papers referring to relations between delusions and physical diseases such as knee osteoarthritis. Even more interesting results were obtained after adding context information. The demographic data show some relation between comorbidity of \(\{ F20, M17\}\) and location of thermal springs in Bulgaria (Fig. 3). Expected BMI values of these patients are high but most of them have normal BMI or a little overweigh. Thus, contextualizing the FPM findings, the proposed technology supports discovery and exploration of novel correlations between phenotypes and comorbidity.
The role of phenotype for comorbidity of various diseases is known. For instance, the most often psychiatric disorder—depression—is comorbid with anxiety disorders, abuse with psychoactive substances, alcohol and drug dependence. High comorbidity is established between depression and somatic dysfunctions as well, e.g. 22–33% of the patients hospitalised for treatment of somatic diseases have depressive disorders too [16]. It is accepted that the predisposition to the development of certain disease is due to the contribution of multiple genes with little effect. The correlation between the genetic fingerprint and the environment works in both directions: people with genetic predisposition can develop certain illness when they live in the respective environment; on the other hand the genes can change the individual sensitivity to the environmental factors and contribute to the development of predisposition [17].
The experiments presented here show that deeper understanding of the interrelations between comorbidity, phenotypes and environmental factors can be achieved by finer tuning of the classical data mining techniques in order to discover unknown correlations between data items in patient records and contextual information.
Conclusion and future work
This paper presents a novel algorithm MixCO for MFI mining. The main advantage of MixCO is that it can process efficiently big dense datasets for small relative minsup values. This is a bottom-up approach which eliminates at the beginning the most critical items that are highly possible to be reduced in the MFI. The expected application impact of MixCO is significant. The explication of maximal frequent itemsets enables to build hypotheses concerning the causality relationships among the exogeneous and endogeneous factors that trigger the formation of these sets. Mining of patterns is shown here, and mining sequences is the next task in our agenda.
Future work includes also in-depth experiments with various OR subsets and evaluation of the effectiveness of MixCO.
The diagnoses with several possible ICD-10 codes or similar diagnoses are also not interpreted in this model. This is an important issue and we plan further investigation of it in our future work.
Finally we note that the technology can be successfully used for explication of risk groups of patients that have predisposition to develop socially-significant disorders like diabetes. This is possible given the large repository of patient-related data organised now in a national Diabetes Register for Bulgaria.Footnote 6 In this way advanced DM algorithms like MixCO and their application to repositories like the Diabetes Register in Bulgaria will turn static archives to powerful alerting and predictive frameworks.
Notes
Clinical Text Analysis and Knowledge Extraction System: http://ctakes.apache.org/.
International Classification of Diseases and Related Health Problems 10th Revision. http://apps.who.int/classifications/icd10/browse/2015/en.
Chronic diseases, WHO, http://www.who.int/topics/chronic_diseases/en/.
WHO, Standard age groups http://www.who.int/healthinfo/paper31.pdf.
WHO, BMI Classification http://apps.who.int/bmi/index.jsp?introPage=intro_3.html.
References
Nasreen S, Azam MA, Shehzad K, Naeem U, Ghazanfar MA. Frequent pattern mining algorithms for finding associated frequent patterns for data streams: a survey. Procedia Comput Sci. 2014;37:109–16.
Agrawal R, Imielinski T, Swami A. Mining association rules between sets of items in large databases. In: Proc. ACM Int. Conf. on management of data (SIGMOD), 1993, p. 207–216.
Dao-I Lin, Kedem. Zvi M.: Pincer search: a new algorithm for discovering the maximum frequent set, in advances in database technology. In: Proc. of the 6th international conference on extending database technology (EDBT’98), 1998, p. 105–119.
Gouda K, Zaki MJ. GenMax: an efficient algorithm for mining maximal frequent itemsets. Data Min Knowl Disc. 2005;11(3):223–42.
Grahne G, Zhu J. Efficiently using prefix-trees in mining frequent itemsets. In: FIMI 90, November 2003.
Burdick D, et al. MAFIA: a maximal frequent itemset algorithm. IEEE Trans Knowl Data Eng. 2005;17(11):1490–504.
Goryachev S, Sordo M, Zeng QT. A suite of natural language processing tools developed for the I2B2 project. In: AMIA annu symp proc, vol. 931, 2006.
Liao KP, Cai T, Savova GK, Murphy SN, Karlson EW, Ananthakrishnan AN, Gainer VS, et al. Development of phenotype algorithms using electronic medical records and incorporating natural language processing. Br Med J. 2015;350(1):h1885.
Xu X, Mishra GD, Jones M. Mapping the global research landscape and knowledge gaps on multimorbidity: a bibliometric study. J Glob Health. 2017;7(1):010414.
Farran B, Channanath AM, Behbehani K, Thanaraj TA. Predictive models to assess risk of type 2 diabetes, hypertension and comorbidity: machine-learning algorithms and validation using national health data from Kuwait–a cohort study. BMJ Open. 2013;3(5):e002457.
Ji X, Chun SA, Geller J. Predicting comorbid conditions and trajectories using social health records. IEEE Trans Nanobiosci. 2016;15(4):371–9.
Boytcheva S. Shallow medication extraction from hospital patient records. In: Koutkias V, Niès J, Jensen S, Maglaveras N, Beuscart R, editors. Studies in health technology and informatics series, vol. 166, IOS Press; 2011. p. 119–128.
Boytcheva S, Angelova G, Angelov Z, Tcharaktchiev D. Text mining and big data analytics for retrospective analysis of clinical texts from outpatient care. Cybern Inf Technol. 2015;15(4):58–77.
Yu H, Hsieh C, Chang K, Lin C. Large linear classification when data cannot fit in memory. ACM Trans Knowl Discov Data (TKDD). 2012;5(4):23.
Neale MC, Kendler KS. Models of comorbidity for multifactorial disorders. Am J Hum Genet. 1995;57:935–53.
Di Matteo MR, Lepper HS, Croghan TW. Depression is a risk factor for noncompliance with medical treatment: meta-analysis of the effects of anxiety and depression on patient adherence. Arch Intern Med. 2000;60(14):2101–7.
Milanova V. Affective disorders: interrelation between genetic and psychosocial factors. Psychosom Med. 2006;14:137–58 (in Bulgarian language).
Acknowledgements
The research work presented in this paper is partially supported by the grant SpecialIZed Data MIning MethoDs Based on Semantic Attributes (IZIDA), funded by the Bulgarian National Science Fund in 2017–2019. The team acknowledges also the support of Medical University—Sofia, the Bulgarian Ministry of Health and the Bulgarian National Health Insurance Fund.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made.
The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder.
To view a copy of this licence, visit https://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Boytcheva, S., Angelova, G., Angelov, Z. et al. Mining comorbidity patterns using retrospective analysis of big collection of outpatient records. Health Inf Sci Syst 5, 3 (2017). https://doi.org/10.1007/s13755-017-0024-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13755-017-0024-y