Abstract
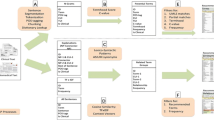
In this paper, we present a case study in using ontologies within a system for identifying patients who are eligible for clinical trials. The main purpose of this clinical research data warehouse (CRDW) is to support patient recruitment based on routine data from the clinical information system. In contrast to most other systems for similar purposes, the CRDW also makes use of information extracted from clinical documents like admission reports, radiological findings and discharge letters. The so-called linguistic pipeline of the CDRW recognizes negated and coordinated phrases. It is supported by clinical application ontologies, which enable the identification of main terms and their properties, as well as semantic search with synonyms, hypernyms, and syntactic variants. In the paper, we discuss questions related to designing the ontologies and filling them with content. The CRDW is currently being tested at several departments of the Charité—Universitätsmedizin Berlin and the Vivantes—Netzwerk für Gesundheit GmbH. In the article, we provide a thorough evaluation of the deployed systems based on real data related to clinical trials conducted by our neurology departments.





Similar content being viewed by others
References
Bodenreider O (2004) The unified medical language system (umls): integrating biomedical terminology. Nucl Acids Res 32:267–270 (Database-Issue)
Brinkman RR, Courtot M, Derom D, Fostel JM, He Y, Lord P, Malone J, Parkinson H, Peters B, Rocca-Serra P et al (2010) Modeling biomedical experimental processes with obi. J Biomed Semant 1(Suppl 1):S7
Broekstra J, Kampman A, Van Harmelen F (2002) Sesame: a generic architecture for storing and querying rdf and rdf schema. In: The Semantic Web-ISWC 2002, Springer, Berlin, p 54–68
Browne P (2009) Jboss drools business rules. From technologies to solutions, Packt Publishing Limited, India, http://books.google.de/books?id=aweMMi7PttwC
Chinchuluun A, Pardalos P, Migdalas A, Pitsoulis L (2008) Pareto optimality, game theory and equilibria, vol 17. Springer, New York
Cowie J, Wilks Y (2000) Information extraction. In: Dale R, Moisl H, Somers H (eds) Handbook of natural language processing. CRC Press, Boca Raton, Florida, USA, pp 241–260
Cunningham H, Tablan V, Roberts A, Bontcheva K (2013) Getting more out of biomedical documents with GATE’s full lifecycle open source text analytics. PLoS Comput Biol 9(2):e1002854 doi:10.1371/journal.pcbi.1002854
Dugas M, Lange M, Berdel W, Müller-Tidow C (2008) Workflow to improve patient recruitment for clinical trials within hospital information systems: a case-study. Trials 9(1):2
Gallaire H, Minker J, Nicolas JM (1984) Logic and databases: a deductive approach. ACM Comput Surv 16(2):153–185
Glock J, Herold R, Pommerening K (2006) Personal identifiers in medical research networks: evaluation of the personal identifier generator in the competence network paediatric oncology and haematology. Submitted for publiction
Graubner B (2007) ICD und OPS. Bundesgesundheitsblatt-Gesundheitsforschung-Gesundheitsschutz 50(7):932–943
Horrocks I, Patel-Schneider PF, Boley H, Tabet S, Grosof B, Dean M (2004) SWRL: a semantic web rule language combining OWL and RuleML, vol 21. W3C Member submission. W3C, p 79. http://www.w3.org/Submission/SWRL/.
Hripcsak G, Ludemann P, Pryor TA, Wigertz OB, Clayton PD (1994) Rationale for the Arden syntax. Comput Biomed Res 27(4):291–324
Hussain S, Ouagne D, Sadou E, Dart T, Jaulent MC, Vloed BD, Colaert D, Daniel C (2012) EHR4CR: a semantic web based interoperability approach for reusing electronic healthcare records in protocol feasibility studies. In: Paschke A, Burger A, Romano P, Marshall MS, Splendiani A (eds) SWAT4LS, CEUR-WS.org, CEUR Workshop Proceedings, vol 952
Jurafsky D, Martin JH (2008) Speech and language processing, 2nd edn., Series in artificial intelligence. Prentice Hall, Upper Saddle River, New Jersey, USA
Kifer M (2008) Rule interchange format: the framework. Web reasoning and rule systems. In: Calvanese D, Lausen G (eds) LNCS, vol 5341. Springer, Berlin Heidelberg, Germany, pp 1–11
Kifer M, Lausen G, Wu J (1995) Logical foundations of object-oriented and frame-based languages. J ACM 42(4):741–843
Lloyd JW (1987) Foundations of logic programming, 2nd edn. Springer-Verlag New York, Inc. New York, USA
Lo O, Fan L, Buchanan WJ, Thuemmler C (2012) Technical evaluation of an e-health platform. In: Macedo M (ed) IADIS E-Health, IADIS, pp 21–28. ISBN 978-972-8939-70-0
Marwede D, Schulz T, Kahn T (2008) Indexing thoracic CT reports using a preliminary version of a standardized radiological lexicon (radlex). J Digit Imag 21(4):363–370
Miles A, Pérez-Agüera JR (2007) SKOS: simple knowledge organisation for the web. Catal Class Quarterly 43(3–4):69–83
Müller F (2005) A finite-state approach to shallow parsing and grammatical functions annotation of german. PhD thesis, University of Tubingen, Tübingen, Germany
Murphy SN, Mendis ME, Berkowitz DA, Chueh IKH (2006) Integration of clinical and genetic data in the i2b2 architecture. In: AMIA Annu Symp Proc, (2009)
Polleres A (2007) From SPARQL to rules (and back). In: Zurko ME, Patel-Schneider PF, Shenoy PJ, Williamson CL (eds.) ACM, USA, p 787–796
Rector A, Rogers J, Zanstra P, Van Der Haring E (2003) OpenGALEN: open source medical terminology and tools. In: AMIA Annual Symposium Proceedings, American Medical Informatics Association, vol 2003, p 982
Reeve L (2005) Survey of semantic annotation platforms. In: Proceedings of the 2005 ACM Symposium on Applied Computing, ACM Press, USA, p 1634–1638
Rogers FB (1963) Medical subject headings. Bull Med Libr Assoc 51:114–116
Rosse C, Mejino J (2003) A reference ontology for biomedical informatics: the foundational model of anatomy. J Biomed Inform 36:478–500
Ruch P, Gobeill J, Lovis C, Geissbuhler A (2008) Automatic medical encoding with SNOMED categories. BMC Med Inform Dec Making 8:6
Russell SJ, Norvig P (2003) Artificial intelligence: a modern approach, 2nd edn. Prentic Hall, Upper Saddle River, New Jersey, USA
Savova GK, Masanz JJ, Ogren PV, Zheng J, Sohn S, Schuler KK, Chute CG (2010) Mayo clinical text analysis and knowledge extraction system (cTAKES): architecture, component evaluation and applications. JAMIA 17(5):507–513
Scheitz JF, Mochmann HC, Nolte CH, Haeusler KG, Audebert HJ, Heuschmann PU, Laufs U, Witzenbichler B, Schultheiss HP, Endres M (2011) Troponin elevation in acute ischemic stroke (TRELAS)—protocol of a prospective observational trial. M BMC Neurol 11:98
Scheitz JF, Mochmann HC, Fiebach BWB, Audebert HJ, Nolte CH (2012) J Neurol 259(1):188–190
Shvaiko P, Euzenat J (2013) Ontology matching: state of the art and future challenges. Knowledge and Data Engineering, IEEE Transactions on , vol 25, issue 1. IEEE Press, Piscataway, NJ, USA
Staab S, Studer R (2009) Handbook on ontologies, 2nd edn. Springer, Berlin Heidelberg, Germany
Szarvas G, Farkas R, Busa-Fekete R (2007) Research paper: state-of-the-art anonymization of medical records using an iterative machine learning framework. JAMIA 14(5):574–580
Todorov K, Geibel P, Kuhnberger KU (2010) Mining concept similarities for heterogeneous ontologies. In: Perner P (ed) Advances in data mining. Applications and theoretical aspects, vol 6171. Springer, Berlin, pp 86–100
Wimalasuriya DC, Dou D (2010) Ontology-based information extraction: an introduction and a survey of current approaches. J Info Sci 36(3):306–323
World Health Organization (2004) ICD-10: International statistical classification of diseases and related health problems. World Health Organization, Geneva, Switzerland
Yu L (2011) A developers guide the semantic web. Springer, Berlin Heidelberg, Germany
Acknowledgments
The project was partially funded by TSB Technologiestiftung Berlin, Zukunftsfonds Berlin, and co-financed by the European Union—European Fund for Regional Development. We would like to thank all computer scientists, linguists, ontologists, physicians, study nurses, and administrative staff of all three partners, who participated in the organization of the project and development of the software.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Geibel, P., Trautwein, M., Erdur, H. et al. Ontology-Based Information Extraction: Identifying Eligible Patients for Clinical Trials in Neurology. J Data Semant 4, 133–147 (2015). https://doi.org/10.1007/s13740-014-0037-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13740-014-0037-5