Abstract
Ruminant enteric methane emission contributes to global warming. Although breeding low methane-emitting cows appears to be possible through genetic selection, doing so requires methane emission quantification by using elaborate instrumentation (respiration chambers, SF6 technique, GreenFeed) not feasible on a large scale. It has been suggested that milk fatty acids are promising markers of methane production. We hypothesized that methane emission can be predicted from the milk fatty acid concentrations determined by mid-infrared spectroscopy, and the integration of energy-corrected milk yield would improve the prediction. Therefore, we examined relationships between methane emission of cows measured in respiration chambers and milk fatty acids, predicted by mid-infrared spectroscopy, to derive diet-specific and general prediction equations based on milk fatty acid concentrations alone and with the additional consideration of energy-corrected milk yield. Cows were fed diets differing in forage type and linseed supplementation to generate a large variation in both CH4 emission and milk fatty acids. Depending on the diet, equations derived from regression analysis explained 61 to 96% of variation of methane emission, implying the potential of milk fatty acid data predicted by mid-infrared spectroscopy as novel proxy for direct methane emission measurements. When data from all diets were analyzed collectively, the equation with energy-corrected milk yield (CH4 (L/day) = − 1364 + 9.58 × energy-corrected milk yield + 18.5 × saturated fatty acids + 32.4 × C18:0) showed an improved coefficient of determination of cross-validation R2CV = 0.72 compared to an equation without energy-corrected milk yield (R2CV = 0.61). Equations developed for diets supplemented by linseed showed a lower R2CV as compared to diets without linseed (0.39 to 0.58 vs. 0.50 to 0.91). We demonstrate for the first time that milk fatty acid concentrations predicted by mid-infrared spectroscopy together with energy-corrected milk yield can be used to estimate enteric methane emission in dairy cows.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Livestock contributes approximately 18% to the global anthropogenic greenhouse gas emission (Hristov et al. 2013). Methane (CH4) mitigation through feeding and genetic selection has been reported to range from 2.5 to 19% in cattle (Knapp et al. 2014; Pickering et al. 2015). Genetic selection of low methane-emitting cows requires quantifying individual methane emission in large populations. However, in vivo methane quantification via respiration chambers, the SF6 technique, and the GreenFeed system are not feasible on a large scale (Hammond et al. 2016). Biochemical relationships among rumen fermentation pathways, CH4 production, and milk fatty acid composition suggest the usefulness of milk fatty acid profiles to predict methane emission (liters/day) (Knapp et al. 2014; van Lingen et al. 2014) (Fig. 1). There have been several reports on the relationship between milk fatty acids and methane emission as well as on equations predicting methane yield defined as liters of CH4 per kilogram dry matter intake and methane intensity defined as liters of CH4 per kilogram energy-corrected milk yield (Castro-Montoya et al. 2017; van Lingen et al. 2014). Besides dry matter intake, the dietary composition, especially forage type and fat supplements, affects milk fatty acid profile and influences methane prediction equations (Dijkstra et al. 2016; Rico et al. 2016; van Lingen et al. 2014). Thus, diet-specific and general equations for estimation of methane emission are important for different farming conditions. Although gas chromatography is the reference technique to measure milk fatty acids, it is unsuitable for routine milk fatty acid determination. In contrast to gas chromatography, mid-infrared spectroscopy can inexpensively predict major milk fatty acids and groups of fatty acids but prediction quality differs among fatty acids (De Marchi et al. 2014; Soyeurt et al. 2011). A Belgian group predicted CH4 emission by directly using the global milk mid-infrared spectra with partial least squares regression and compared results with multiple correlations between milk fatty acids and CH4 (Dehareng et al. 2012). These authors concluded that global milk mid-infrared spectra could be more feasible than predicted milk fatty acids to estimate CH4 emission (Dehareng et al. 2012). The methane prediction derived from global mid-infrared milk spectra by Dehareng et al. (2012) was further improved by including information on lactation stage (Vanlierde et al. 2015; Vanlierde et al. 2016). Dry matter intake is the main determinant for CH4 emission (Hristov et al. 2013; Knapp et al. 2014), but individual cow intake information is not available on farms. In contrast, individual records of energy-corrected milk yield, which is highly related to dry matter intake (Hristov et al. 2013), can readily be calculated from milk testing data. Thus, we hypothesized that CH4 emission can be predicted from milk fatty acid concentrations determined by mid-infrared spectroscopy, and that the integration of energy-corrected milk yield improves prediction. Our objective was to develop equations that predict CH4 emission on the basis of milk fatty acid composition estimated by mid-infrared spectroscopy. We further investigated whether inclusion of energy-corrected milk yield, compared with dry matter intake, could improve the prediction. The datasets were based on CH4 emission of dairy cows measured in respiration chambers fed diets differing in forage type and linseed supplementation to generate a larger variation in both CH4 emission and milk fatty acids.
2 Materials and methods
2.1 Animals, experimental design, and diets
Ethical approval for the study was obtained (permission no. 7221.3-1-014/14). Twenty half-sib German Holstein cows (106 ± 28 days in milk, 580 ± 57 kg bodyweight, mean ± standard deviation) in second (15 cows) or third lactation (5 cows) were used. The required number of cows for this study was determined through a power test for crossover experimental designs as described (Rasch et al. 1998) using CADEMO software (Windows ANOV-Version 4.03 (2000); BioMath GmbH, Rostock, Germany). Cows were kept in tie stalls and provided with water and total mixed rations for ad libitum intake. Two isoenergetic total mixed rations, in which the major forage component was either corn silage or grass silage both with or without linseed supplementation, were used (7 MJ NEL/kg dry matter) to generate a wide range of variation in both methane emissions and milk fatty acid profiles. Diets contained grass and corn silage at dry matter levels of 130 and 450 g/kg (corn silage-based diets) and 360 and 190 g/kg (grass silage-based diets), respectively. Linseed-supplemented diets contained 60 g of fat/kg dry matter compared to 30 g of fat in diets without supplementation. The target nutrient content of the diets was according to recommendations of the German Society of Nutritional Physiology (Gesellschaft für Ernährungsphysiologie 2001). Randomly selected cows (n = 10) were fed corn silage-based diets for 5 weeks without linseed supplementation and subsequently for another 5 weeks with linseed supplementation with 1 week in-between for a stepwise change of diets (11 experimental weeks per cow in total) in a randomized order (5 cows started with corn silage without linseed). The other 10 cows were allocated randomly to grass silage diets without or with linseed supplementation for 5 weeks each (5 cows started with grass silage without linseed). Feed intake was recorded daily. Residual feed was removed from the trough before cows were offered fresh feed at 0730 and 1730 h. Cows were milked at 0630 and 1630 h. Three cows feeding on the grass silage-based diet supplemented with linseed had to be excluded from analysis because of illness or feed refusal.
2.2 Determination of methane production and feed and milk composition
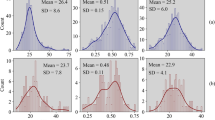
Methane emission of each cow was quantified in separate respiration chambers in experimental weeks 5 and 11. Cows entered the chambers 15 h before starting two subsequent 24-h CH4 measurements (0700 to 0700 h). The average 24-h CH4 emission was calculated for each cow (Table 1). In the chambers, the temperature and relative humidity were 15 °C and 60%, respectively, and the light was on from 0600 to 1900 h (Derno et al. 2009). The CH4 recovery in the chambers was 99.9%. Analyses of feed were performed according to the Weender standard analysis (Naumann et al. 1976) with modifications by van Soest et al. (1991) by the accredited feed laboratory Landwirtschaftliche Untersuchungs- und Forschungsanstalt der LMS Agrarberatung GmbH (LUFA, Rostock, Germany). The energy content was calculated as recommended (Gesellschaft für Ernährungsphysiologie 2001). During the respiration measurements, milk aliquots of one evening and the following morning milking were pooled (0.25% of milk yield per milking). Milk fat and protein content was analyzed using mid-infrared spectroscopy (MilkoScan FT6000 and MilkoScan FT+; Foss, Hilleroed, Denmark) by the state control association Mecklenburg-Western Pomerania (Landeskontrollverband für Leistungs- und Qualitätsprüfung Mecklenburg-Vorpommern e.V., Güstrow, Germany). Energy-corrected milk yield (kg/d) was calculated as [((0.38 × milk fat %) + (0.21 × milk protein %) + 1.05)/3.28 × milk yield (kg/d)] (Spiekers et al. (2009). Milk fat and fatty acid compositions were analyzed using mid-infrared spectroscopy (MilkoScan FT6000) by a commercial laboratory (Comité du Lait, Battice, Belgium). Multivariate calibration equations using partial least squares regression were used to derive milk fatty acid concentrations. One milk sample (grass silage without linseed group) produced invalid results which were not used in the statistical analysis. Contents of saturated and unsaturated fatty acids, as well as isomers of C18:1cis and trans and ω-3 fatty acids, were predicted as groups and were expressed as the percentage of total milk fat. Mono- and polyunsaturated fatty acids were expressed as the percentage of unsaturated fatty acids in milk fat. Milk palmitic (C16:0) and stearic acids (C18:0) were estimated individually. Performance data of milk fatty acid prediction are given in Table 1.
2.3 Statistical analyses
2.3.1 Development of prediction equations and computing of correlations
Methane emission and CH4 intensity were estimated using multiple linear regression analysis with the stepwise explanatory-independent variable selection method for each diet separately. Methane emission was also predicted based on combined data categorized by basal diet or linseed supplementation, as well as for all available data across four diets to explore diet dependency of methane prediction equations. This was done to test different feeding scenarios and to explore how explanatory milk fatty acids differ in prediction models for various diets. For the regression analysis, three datasets with different independent explanatory variables were made available. The first dataset included only the milk fatty acid concentrations (nine items: groups or individual milk fatty acids; Table 1), whereas the second set additionally included the energy-corrected milk yield, and the third set included dry matter intake instead of energy-corrected milk yield. Prediction models for CH4 intensity were generated with datasets 1 and 3. The entry and stay levels of explanatory variables for the stepwise (forward) variable selection method were set at P ≤ 0.15. Selection of best models was based on improvement in the multiple fit parameters as described by Kaps and Lamberson (2004). According to this, we selected best models that simultaneously maximized the coefficient of determination (R2Model) and minimized the conceptual predictive criterion (Cp), implying that the lowest number of variables that could explain the highest variation was allowed to enter the equations. In a separate crosscheck, it was also ensured that best model selection based on the two fit parameters (R2Model and Cp) also corresponded to the lowest Akaike’s Information Criteria and residual sum of squares, two other commonly used fit parameters. For the sake of clarity, we however report only R2Model as a fit parameter because it is most commonly used and corresponds to the proportion of variation explained. Using the predicted milk fatty acid and measured methane emission data, Pearson correlations were computed. A Spearman rank correlation analysis was also performed to quantify whether the cow rankings for CH4 emission levels were consistent between consecutive periods, when cows were fed diets with or without linseed in two different periods.
2.3.2 Cross-validation of prediction equations
To assess the predictive ability of the developed methane prediction models, an internal cross-validation was performed using SAS software (SAS Institute Inc. 2017). The cross-validation was based on the above-described datasets and run through a stepwise approach. Data sub-sets were generated by omitting data of one animal at a step, and the emission of the omitted animal was predicted using the regression function with the data from remaining animals as described (Moraes et al. 2014). Thus, methane prediction for a specific animal was generated using models fitted without the observation of these specific animals. The root mean square error of cross-validation (RMSE) and the cross-validation coefficient of determination (R2CV) were then estimated to evaluate the goodness of model fit for the cross-validation.
3 Results and discussion
3.1 Methane emission and milk fatty acid composition
The dataset included a total of 36 observations from 20 cows to develop methane prediction equations feeding four different diets varying in basal rations (grass silage or corn silage) with or without linseed supplementation. The diets provoked a wide range of methane yield with a minimum of 21 and a maximum of 50 L/kg dry matter intake, and milk fatty acid composition ranged from 55 to 78% saturated fatty acids of total fat (Table 1). Results of studies in which 30 different diets with and without lipid supplements were fed were used in a meta-analysis (van Lingen et al. 2014); the methane yield (21.5 ± 2.46 g/kg of dry matter intake; mean ± SD) was lower than that observed in our study but milk fat levels of C16:0 (31.3 ± 4.91 g/100 g of total milk fat) and C18:0 (9.8 ± 2.41 g/100 g of total milk fat) were comparable (Table 1). When diets based on grass and corn silage were fed, mean methane yields (24.1 ± 1.87 g/kg of dry matter intake) and milk fat levels of C16:0 (35.4 ± 2.72 g/100 g of total fatty acids) and C18:0 (7.5 ± 1.03 g/100 g of total fatty acids) were reported (van Gastelen et al. 2017) but variability was smaller than in our and the van Lingen et al. (2014) study.
Our results for milk fatty acid composition were largely in line with an earlier study where milk fatty acid contents were also determined by mid-infrared spectroscopy-based predictions (Soyeurt et al. 2011). It has been shown that milk fatty acid composition analyzed by mid-infrared spectroscopy as compared to gas chromatography corresponded well (Soyeurt et al. 2011). Predictions of milk fatty acid contents by mid-infrared spectroscopy validated by gas chromatography showed high accuracies (R2CV ≥ 0.88) for individual saturated fatty acids, C16:0, C18:0, C18:1trans, and C18:1cis, and groups of saturated, unsaturated, and monounsaturated fatty acids, whereas accuracy was R2CV = 0.75 for ω-3 fatty acids, which was similar in our study (Table 1). This agreement between mid-infrared spectroscopy and gas chromatography is the prerequisite to use mid-infrared spectroscopy for routine milk fatty acid recordings.
Furthermore, we calculated rank correlation coefficients of r = 0.78 and r = 0.73 (P = 0.001) for CH4 emission and CH4 yield, respectively (Fig. 2). These results indicate replicable responses under the effects of either diet with or without linseed, representing different environmental conditions, and suggested the existence of a genetic component responsible for the individual methane production level.
Correlation of measured CH4 emission in liters per day and measured CH4 emission per kilogram dry matter intake between linseed supplemented and non-supplemented diets considering individual animals (A, B, C, …), and basal ration (grass silage = green, corn silage = orange) indicating repeatable phenotypes under the effect of either diets with or without linseed. The shaded area depicts the confidence ellipse. The Spearman rank correlation coefficient (r) and the probability (p) are given
3.2 Predicting methane emission
Most studies reporting prediction equations have been based on milk fatty acid concentrations analyzed by gas chromatography (Castro-Montoya et al. 2017; Rico et al. 2016; van Lingen et al. 2014). Published prediction models based on global infrared milk spectra have used the model of Dehareng et al. (2012) or further improvement of the model by integrating the lactation stage (Vanlierde et al. 2015; Vanlierde et al. 2016). Dehareng et al. (2012) included methane production data of 11 cows and have reported R2CV between 0.68 and 0.79 for grams of CH4 per day and grams of CH4 per kilogram of milk, respectively. Shetty et al. (2017) developed methane prediction models based on global infrared milk spectra with R2Validation = 0.13 and concluded that it is not feasible to predict methane emission (liters/day) based on mid-infrared spectra alone. Upon integrating milk yield, the prediction accuracy was improved (R2Validation = 0.35) (Shetty et al. 2017). In the present study, three datasets were used to develop multiple regression equations. When energy-corrected milk yield was additionally included (dataset 2), the goodness of fit was improved by up to 25% relative to the equations based on dataset 1 (Table 2). When energy-corrected milk yield was replaced by dry matter intake in the model (dataset 3; Table 2), the goodness of fit was improved by up to 41% relative to equations derived from dataset 1. Although dry matter intake explains a large part of the variation observed in methane emissions (Knapp et al. 2014), this information is not available on farms. Thus, it cannot be considered as a variable to be implemented in methane prediction equations for on-farm applications. In contrast, energy-corrected milk yield is a readily available cow individual measure reflecting the dry matter intake (Hristov et al. 2013) and consequently can be used as a surrogate for dry matter intake. Using the first dataset, the goodness of the regression fit for CH4 emission for individual diets ranged from R2Model = 0.61 to 0.94 (Table 2). Using data from all four diets collectively, a coefficient of determination of R2Model = 0.70 was obtained.
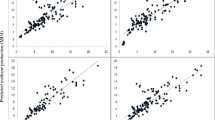
The equations based on datasets 1 to 3 were validated by an internal cross-validation (Table 2). The R2CV ranged between 0.39 and 0.91 with a RMSE between 147 and 35 liters/day and the R2CV was lowest for linseed-supplemented diets. If linseed-supplemented diets were not considered, the R2CV was never lower than 0.50. When coefficients of determination were compared between dataset 1 and datasets 2 and 3 for all four diets collectively, the R2CV was improved from 0.61 to 0.72 by the integration of energy-corrected milk yield (Fig. 3), and to R2CV = 0.75 by the integration of dry matter intake as explanatory variables. In previous studies predicting CH4 emission (g/day) using gas chromatography to determine milk fatty acid concentrations, improvements of goodness of fit values by 5 to 12% were achieved when dry matter intake was considered in the model (Mohammed et al. 2011; Rico et al. 2016). In a meta-analysis, improved indices of predictive ability were observed when dry matter intake was included (Castro-Montoya et al. (2017). Nevertheless, a further cross-validation of our prediction equation using an independent validation set of cows fed different diets is desirable.
Measured and predicted CH4 emission derived from the cross-validation of multiple regression equations across all experimental diets (grass silage without linseed, corn silage without linseed, grass silage with linseed and corn silage with linseed) based on dataset 1 (milk fatty acids only) and dataset 2 (milk fatty acids plus energy-corrected milk yield). The shaded area depicts the confidence ellipse. The cross-validation coefficient of determination (R2Cross-validation) and the root mean square error (RMSE) of cross-validation are given
A further factor which might be important for methane prediction is stage of lactation, because in early lactation, fatty acids entering milk fat are derived to a larger degree from lipolysis of lipid resources of cows, which can affect explanatory fatty acids in methane prediction models (Vanlierde et al. 2015). The patterns of fatty acids from endogenous lipid depots differ from fatty acids in plant oils (Bayat et al. 2018). Vanlierde et al. (2015) tested the effect of inclusion of days in milk in the prediction model based on milk mid-infrared spectra and showed that lactation stage-dependent prediction is more meaningful compared to lactation stage-independent prediction of methane emission although coefficient of determination did not improve. In a recent study, integrating lactation stage in the model did not improve methane predictive ability (R2Validation = 0.14 vs. 0.13) (Shetty et al. 2017). In our study, all cows were in the same lactation stage which might be limiting the validity of our equation to cows in established lactation. This suggests that further research is required on the effect of lactation stage for milk fatty acid-based methane prediction.
Dietary intake of ω-3 fatty acids with linseed reduces methane emissions and increases the proportion of polyunsaturated fatty acids in milk whereas it can inhibit mammary lipid synthesis by fatty acid intermediates produced during ruminal bio-hydrogenation (Bauman et al. 2011; Bayat et al. 2018). This complex relationship might play a role in the variability of methane production and milk fatty acid contents. Notably, the predictions obtained in this study with linseed-supplemented diets showed smaller goodness of fit values and cross-validation coefficient of determination values (Table 2) although the RMSE was highest for L0 diets in dataset 1. However, when energy-corrected milk yield or dry matter intake was considered as additional explanatory variable (datasets 2 and 3), also in diets with linseed a larger part of variation could be explained. This implies that inclusion of other parameters explaining additional variation of methane emission should also be considered.
Prediction of CH4 intensity would be desirable because it considers the product level. However, in our study, CH4 intensity was not a robust parameter; prediction equations using the first dataset produced largely insignificant weak to moderate fit values. The use of dataset 3 did not generate considerable improvements when compared to dataset 1. In line with this, others found moderate to low R2 of 0.47 and R2Validation of 0.18 (Castro-Montoya et al. 2017; van Lingen et al. 2014) for predicted CH4 emission per kilogram milk based on milk fatty acid data only.
The explanatory fatty acids in published prediction equations for methane emission differ considerably between studies (Castro-Montoya et al. 2017; van Lingen et al. 2014). The milk fatty acids C16:0 and C18:1cis and trans isomers are frequently found in prediction models for CH4 emission (Castro-Montoya et al. 2017; Chilliard et al. 2009; Rico et al. 2016). A positive relationship with methane emission was found for C16:0 in this (r = 0.57, P < 0.001) and other studies (Castro-Montoya et al. 2017; Chilliard et al. 2009; van Lingen et al. 2014). Palmitic acid is the saturated fatty acid with the highest concentration in plant feed and is also synthesized de novo from acetate in animal tissues, and acetate concentration in the rumen is directly associated with methane production (van Lingen et al. 2014). In contrast, C18:0 (r = − 0.34, P = 0.044) and C18:1cis isomers (r = − 0.61, P < 0.001) were negatively related to methane emission in our study. Vanrobays et al. (2016) have found weak negative relations for C18:0 and C18:1cis9 analyzed with mid-infrared spectroscopy, whereas studies using gas chromatography have reported no correlation for C18:0, but various C18:1cis isomers, and the sum of C18 fatty acids was negatively related to methane production (Chilliard et al. 2009; Mohammed et al. 2011; Rico et al. 2016). Stearic acid is an end product of C18:3 ω-3 bio-hydrogenation and C18:3 ω-3 is particularly abundant in linseed. Products of bio-hydrogenation and intermediates were negatively related to CH4 emission (van Gastelen and Dijkstra 2016). In our study, C18:1trans isomers were a factor in the model for CH4 emission in the equation of dataset 1 for all diets. They tended to be negatively related to methane emission (r = − 0.31, P = 0.066), in line with findings from other studies (Mohammed et al. 2011; Rico et al. 2016). Groups of fatty acids appeared to play a minor role as prediction variables in our models, but the group of saturated fatty acids was positively associated with methane emission (r = 0.60, P < 0.001) as was also found by others (van Gastelen and Dijkstra 2016; Vanrobays et al. 2016). No specific pattern for milk fatty acids as explanatory variables could be identified in the equations. However, it appears that among the milk fatty acids considered in the datasets, C16:0 plays a dominant role in the prediction of methane emission by milk fatty acids.
4 Conclusion
The prediction of methane emission using data of all four diets collectively showed moderate predictive accuracies (R2CV between 0.61 and 0.75). When energy-corrected milk yield or dry matter intake was additionally considered in the models, prediction accuracy improved by 18 and 23%, respectively. Thus, energy-corrected milk yield in prediction equations is a useful replacement for dry matter intake which cannot be determined on farm. Models developed for diets including linseed seem to be lower in prediction accuracy which deserves further research. We conclude that methane prediction equations based on milk fatty acids estimated by mid-infrared spectroscopy and energy-corrected milk yield could be used as a screening method for individual methane emission of cows.
References
Bauman DE, Harvatine KJ, Lock AL (2011) Nutrigenomics, rumen-derived bioactive fatty acids, and the regulation of milk fat synthesis. Annu Rev Nutr 31:299–319. https://doi.org/10.1146/annurev.nutr.012809.104648
Bayat AR, Tapio I, Vilkki J, Shingfield KJ, Leskinen H (2018) Plant oil supplements reduce methane emissions and improve milk fatty acid composition in dairy cows fed grass silage-based diets without affecting milk yield. J Dairy Sci 101:1136–1151. https://doi.org/10.3168/jds.2017-13545
Castro-Montoya JM, Peiren N, Veneman J, De Baets B, De Campeneere S, Fievez V (2017) Predictions of methane emission levels and categories based on milk fatty acid profiles from dairy cows. Animal 11:1153–1162. https://doi.org/10.1017/S1751731116002627
Chilliard Y, Martin C, Rouel J, Doreau M (2009) Milk fatty acids in dairy cows fed whole crude linseed, extruded linseed, or linseed oil, and their relationship with methane output. J Dairy Sci 92:5199–5211. https://doi.org/10.3168/jds.2009-2375
De Marchi M, Toffanin V, Cassandro M, Penasa M (2014) Invited review: mid-infrared spectroscopy as phenotyping tool for milk traits. J Dairy Sci 97:1171–1186. https://doi.org/10.3168/jds.2013-6799
Dehareng F, Delfosse C, Froidmont E, Soyeurt H, Martin C, Gengler N, Vanlierde A, Dardenne P (2012) Potential use of milk mid-infrared spectra to predict individual methane emission of dairy cows. Animal 6:1694–1701. https://doi.org/10.1017/S1751731112000456
Derno M, Elsner HG, Paetow EA, Scholze H, Schweigel M (2009) Technical note: a new facility for continuous respiration measurements in lactating cows. J Dairy Sci 92:2804–2808. https://doi.org/10.3168/jds.2008-1839
Dijkstra J, van Gastelen S, Antunes-Fernandes EC, Warner D, Hatew B, Klop G, Podesta SC, van Lingen HJ, Hettinga KA, Bannink A (2016) Relationships between milk fatty acid profiles and enteric methane production in dairy cattle fed grass- or grass silage-based diets. Anim Prod Sci 56:541–548. https://doi.org/10.1071/an15509
Gesellschaft für Ernährungsphysiologie (2001) Empfehlungen zur Energie- und Nährstoffversorgung der Milchkühe und Aufzuchtrinder (recommended energy and nutrient supply for dairy cows and growing cattle). German Society of Nutrition Physiology/ Ausschuss für Bedarfsnormen, No. 8 DLG-Verlag, Frankfurt am Main, Germany
Hammond K, Crompton LA, Bannink A, Dijkstra J, Yanez-Ruiz DR, O'Kiely P et al (2016) Review of current in vivo measurement techniques for quantifying enteric methane emission from ruminants. Anim Feed Sci Technol 219:13–30. https://doi.org/10.1016/j.anifeedsci.2016.05.018
Hristov AN, Oh J, Lee C, Meinen R, Montes F, Ott T et al (2013) Mitigation of greenhouse gas emissions in livestock production: a review of technical options for non-CO2 emissions. Food and Agriculture Organization of the United Nations (FAO), Rome
Kaps M, Lamberson W (2004) Biostatistics for animal science. CABI Publishing, United Kingdom. https://doi.org/10.1079/9780851998206.0000
Knapp JR, Laur GL, Vadas PA, Weiss WP, Tricarico JM (2014) Invited review: enteric methane in dairy cattle production: quantifying the opportunities and impact of reducing emissions. J Dairy Sci 97:3231–3261. https://doi.org/10.3168/jds.2013-7234
Mohammed R, McGinn SM, Beauchemin KA (2011) Prediction of enteric methane output from milk fatty acid concentrations and rumen fermentation parameters in dairy cows fed sunflower, flax, or canola seeds. J Dairy Sci 94:6057–6068. https://doi.org/10.3168/jds.2011-4369
Moraes LE, Strathe AB, Fadel JG, Casper DP, Kebreab E (2014) Prediction of enteric methane emissions from cattle. Glob Chang Biol 20:2140–2148. https://doi.org/10.1111/gcb.12471
Naumann C, Bassler R, Seibold R, Barth C (1976) Methodenbuch III: die chemische Untersuchung von Futtermitteln/ method book III: chemical analysis of feedstuffs. VDLUFA - Verlag, Darmstadt
Pickering NK, Oddy VH, Basarab J, Cammack K, Hayes B, Hegarty RS, Lassen J, McEwan JC, Miller S, Pinares-Patiño CS, de Haas Y (2015) Animal board invited review: genetic possibilities to reduce enteric methane emissions from ruminants. Animal 9:1431–1440. https://doi.org/10.1017/s1751731115000968
Rasch D, Herrendörfer G, Bock J, Victor N, Guiard V (1998) Verfahrensbibliothek: Versuchsplanung und –auswertung - band III (process library: designing experimental methods and evaluating the results—volume III). Oldenbourg Verlag, München
Rico DE, Chouinard PY, Hassanat F, Benchaar C, Gervais R (2016) Prediction of enteric methane emissions from Holstein dairy cows fed various forage sources. Animal 10:203–211. https://doi.org/10.1017/s1751731115001949
SAS Institute Inc (2017) SAS OnlineDoc® Version 9.4. SAS Institute Inc., Cary
Shetty N, Difford G, Lassen J, Lovendahl P, Buitenhuis AJ (2017) Predicting methane emissions of lactating Danish Holstein cows using Fourier transform mid-infrared spectroscopy of milk. J Dairy Sci 100:9052–9060. https://doi.org/10.3168/jds.2017-13014
Soyeurt H, Dehareng F, Gengler N, McParland S, Wall E, Berry DP, Coffey M, Dardenne P (2011) Mid-infrared prediction of bovine milk fatty acids across multiple breeds, production systems, and countries. J Dairy Sci 94:1657–1667. https://doi.org/10.3168/jds.2010-3408
Spiekers H, Nußbaum H, Potthast V (2009) Erfolgreiche Milchviehfütterung/Successful feeding of dairy cattle. DLG-Verlag, Frankfurt am Main
van Gastelen S, Antunes-Fernandes EC, Hettinga KA, Dijkstra J (2017) Relationships between methane emission of Holstein Friesian dairy cows and fatty acids, volatile metabolites and non-volatile metabolites in milk. Animal 11:1539–1548. https://doi.org/10.1017/s1751731117000295
van Gastelen S, Dijkstra J (2016) Prediction of methane emission from lactating dairy cows using milk fatty acids and mid-infrared spectroscopy. J Sci Food Agric 96:3963–3968. https://doi.org/10.1002/jsfa.7718
van Lingen HJ, Crompton LA, Hendriks WH, Reynolds CK, Dijkstra J (2014) Meta-analysis of relationships between enteric methane yield and milk fatty acid profile in dairy cattle. J Dairy Sci 97:7115–7132. https://doi.org/10.3168/jds.2014-8268
van Soest PJ, Robertson JB, Lewis BA (1991) Methods for dietary fiber, neutral detergent fiber, and nonstarch polysaccharides in relation to animal nutrition. J Dairy Sci 74:3583–3597. https://doi.org/10.3168/jds.S0022-0302(91)78551-2
Vanlierde A, Vanrobays ML, Dehareng F, Froidmont E, Soyeurt H, McParland S, Lewis E, Deighton MH, Grandl F, Kreuzer M, Gredler B, Dardenne P, Gengler N (2015) Hot topic: innovative lactation-stage-dependent prediction of methane emissions from milk mid-infrared spectra. J Dairy Sci 98:5740–5747. https://doi.org/10.3168/jds.2014-8436
Vanlierde A, Vanrobays ML, Gengler N, Dardenne P, Froidmont E, Soyeurt H et al (2016) Milk mid-infrared spectra enable prediction of lactation-stage-dependent methane emissions of dairy cattle within routine population-scale milk recording schemes. Anim Prod Sci 56:258–264. https://doi.org/10.1071/an15590
Vanrobays ML, Bastin C, Vandenplas J, Hammami H, Soyeurt H, Vanlierde A, Dehareng F, Froidmont E, Gengler N (2016) Changes throughout lactation in phenotypic and genetic correlations between methane emissions and milk fatty acid contents predicted from milk mid-infrared spectra. J Dairy Sci 99:7247–7260. https://doi.org/10.3168/jds.2015-10646
Acknowledgements
The authors express their gratitude to the teams at the “Tiertechnikum” and “EAR” of the Leibniz Institute for Farm Animal Biology (FBN) for excellent technical assistance. We appreciate the help of Dr. O. Bellmann, institute veterinarian of FBN, with medical care. The selection of cows by F. Schultz (RinderAllianz GmbH, Woldegk, Germany) is gratefully acknowledged. We thank DANONE GmbH, Haar, Germany, for providing milk composition data, and Ms. Krüger and Ms. Göschl of DANONE GmbH for fruitful discussions.
Funding
This work is part of the project “Innovation potential to reduce greenhouse gas emissions in the dairy supply chain” (INNO MilCH4) and was supported by funds from the Federal Ministry of Food and Agriculture based on a decision of the Parliament of the Federal Republic of Germany via the Federal Office for Agriculture and Food under the innovation support program (grant no. 2817501011). The publication of this article was funded by the Open Access Fund of the Leibniz Institute for Farm Animal Biology (FBN), Dummerstorf, Germany.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical approval for the study was obtained (permission no. 7221.3-1-014/14).
Additional information
Patent
A request for grant of a European patent was submitted (application no. EP17189083.3).
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Engelke, S.W., Daş, G., Derno, M. et al. Milk fatty acids estimated by mid-infrared spectroscopy and milk yield can predict methane emissions in dairy cows. Agron. Sustain. Dev. 38, 27 (2018). https://doi.org/10.1007/s13593-018-0502-x
Accepted:
Published:
DOI: https://doi.org/10.1007/s13593-018-0502-x