Abstract
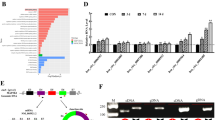
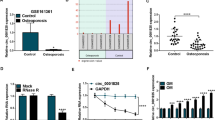
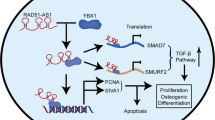
Osteoporosis is a highly prevalent disease characterized by bone mass loss and structural deterioration. There are evidences that altered differentiation of human bone marrow mesenchymal stromal/stem cells (hBMSCs) is a major cause for osteoporosis. Recent studies suggest that circular RNAs (circRNAs) are dysregulated in osteoporosis patients and involved in the pathogenesis of osteoporosis. In the present study, we are aimed to analyze the circRNA expression profiles in osteoporosis patients and identify potential circRNAs that involved in the differentiation of hBMSCs during osteoporosis. Transcriptome RNA-sequencing was conducted to search for differentially expressed circRNAs. Transwell assay, ARS and ALP staining, and ectopic bone formation model were performed to evaluate osteogenic differentiation of hBMSCs. RNA pull-down assay, RNA immunoprecipitation, western blot, and in vitro binding assay were conducted to evaluate the interaction of circRNAs and RNA-binding protein HuR. We found that hsa_circ_0008842 (designated as circZNF367) was upregulated in osteoporosis patients and decreased in hBMSCs during osteogenic differentiation. CircZNF367 overexpression suppressed migration, invasion and osteogenic differentiation of hBMSCs in vitro and in vivo. In comparison, knockdown of circZNF367 promoted migration, invasion and osteogenic differentiation of hBMSCs. CircZNF367 could interact with the RNA-binding protein HuR, thus reduced the mRNA stability of LRP5. Furthermore, HuR overexpression or LRP5 restoration abrogated the effects of circZNF367 overexpression on osteogenic differentiation of hBMSCs. Our results indicated that circZNF367 played a role in osteogenic differentiation of hBMSCs via reducing HuR-mediated mRNA stability of LRP5.







Similar content being viewed by others
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Nih Consensus Development Panel on Osteoporosis Prevention D Therapy. Osteoporosis prevention, diagnosis, and therapy. JAMA. 2001;285(6):785–95. https://doi.org/10.1001/jama.285.6.785.
Sozen T, Ozisik L, Basaran NC. An overview and management of osteoporosis. Eur J Rheumatol. 2017;4(1):46–56. https://doi.org/10.5152/eurjrheum.2016.048.
Morin S, Lix LM, Azimaee M, Metge C, Caetano P, Leslie WD. Mortality rates after incident non-traumatic fractures in older men and women. Osteoporos Int. 2011;22(9):2439–48. https://doi.org/10.1007/s00198-010-1480-2.
Center JR, Nguyen TV, Schneider D, Sambrook PN, Eisman JA. Mortality after all major types of osteoporotic fracture in men and women: an observational study. Lancet. 1999;353(9156):878–82. https://doi.org/10.1016/S0140-6736(98)09075-8.
Tabatabaei-Malazy O, Salari P, Khashayar P, Larijani B. New horizons in treatment of osteoporosis. Daru. 2017;25(1):2. https://doi.org/10.1186/s40199-017-0167-z.
Boyle WJ, Simonet WS, Lacey DL. Osteoclast differentiation and activation. Nature. 2003;423(6937):337–42. https://doi.org/10.1038/nature01658.
Zhou S, Greenberger JS, Epperly MW, Goff JP, Adler C, Leboff MS, Glowacki J. Age-related intrinsic changes in human bone-marrow-derived mesenchymal stem cells and their differentiation to osteoblasts. Aging Cell. 2008;7(3):335–43. https://doi.org/10.1111/j.1474-9726.2008.00377.x.
Shen W, Chen J, Gantz M, Punyanitya M, Heymsfield SB, Gallagher D, Albu J, Engelson E, Kotler D, Pi-Sunyer X, Gilsanz V. MRI-measured pelvic bone marrow adipose tissue is inversely related to DXA-measured bone mineral in younger and older adults. Eur J Clin Nutr. 2012;66(9):983–8. https://doi.org/10.1038/ejcn.2012.35.
Moerman EJ, Teng K, Lipschitz DA, Lecka-Czernik B. Aging activates adipogenic and suppresses osteogenic programs in mesenchymal marrow stroma/stem cells: the role of PPAR-gamma2 transcription factor and TGF-beta/BMP signaling pathways. Aging Cell. 2004;3(6):379–89. https://doi.org/10.1111/j.1474-9728.2004.00127.x.
Chen Q, Shou P, Zheng C, Jiang M, Cao G, Yang Q, Cao J, Xie N, Velletri T, Zhang X, Xu C, Zhang L, Yang H, Hou J, Wang Y, Shi Y. Fate decision of mesenchymal stem cells: adipocytes or osteoblasts? Cell Death Differ. 2016;23(7):1128–39. https://doi.org/10.1038/cdd.2015.168.
Hartmann C. A Wnt canon orchestrating osteoblastogenesis. Trends Cell Biol. 2006;16(3):151–8. https://doi.org/10.1016/j.tcb.2006.01.001.
Yuan Z, Li Q, Luo S, Liu Z, Luo D, Zhang B, Zhang D, Rao P, Xiao J. PPARgamma and Wnt signaling in adipogenic and osteogenic differentiation of mesenchymal stem cells. Curr Stem Cell Res Ther. 2016;11(3):216–25. https://doi.org/10.2174/1574888x10666150519093429.
MacDonald BT, He X. Frizzled and LRP5/6 receptors for Wnt/beta-catenin signaling. Cold Spring Harb Perspect Biol. 2012. https://doi.org/10.1101/cshperspect.a007880.
van Meurs JB, Trikalinos TA, Ralston SH, Balcells S, Brandi ML, Brixen K, Kiel DP, Langdahl BL, Lips P, Ljunggren O, Lorenc R, Obermayer-Pietsch B, Ohlsson C, Pettersson U, Reid DM, Rousseau F, Scollen S, Van Hul W, Agueda L, Akesson K, Benevolenskaya LI, Ferrari SL, Hallmans G, Hofman A, Husted LB, Kruk M, Kaptoge S, Karasik D, Karlsson MK, Lorentzon M, Masi L, McGuigan FE, Mellstrom D, Mosekilde L, Nogues X, Pols HA, Reeve J, Renner W, Rivadeneira F, van Schoor NM, Weber K, Ioannidis JP, Uitterlinden AG, Study G. Large-scale analysis of association between LRP5 and LRP6 variants and osteoporosis. JAMA. 2008;299(11):1277–90. https://doi.org/10.1001/jama.299.11.1277.
Shao T, Pan YH, Xiong XD. Circular RNA: an important player with multiple facets to regulate its parental gene expression. Mol Therapy Nucleic acids. 2021;23:369–76. https://doi.org/10.1016/j.omtn.2020.11.008.
Zhou WY, Cai ZR, Liu J, Wang DS, Ju HQ, Xu RH. Circular RNA: metabolism, functions and interactions with proteins. Mol Cancer. 2020;19(1):172. https://doi.org/10.1186/s12943-020-01286-3.
Zang J, Lu D, Xu A. The interaction of circRNAs and RNA binding proteins: An important part of circRNA maintenance and function. J Neurosci Res. 2020;98(1):87–97. https://doi.org/10.1002/jnr.24356.
Liu H, Lan T, Li H, Xu L, Chen X, Liao H, Chen X, Du J, Cai Y, Wang J, Li X, Huang J, Yuan K, Zeng Y. Circular RNA circDLC1 inhibits MMP1-mediated liver cancer progression via interaction with HuR. Theranostics. 2021;11(3):1396–411. https://doi.org/10.7150/thno.53227.
Wang Y, Liu J, Ma J, Sun T, Zhou Q, Wang W, Wang G, Wu P, Wang H, Jiang L, Yuan W, Sun Z, Ming L. Exosomal circRNAs: biogenesis, effect and application in human diseases. Mol Cancer. 2019;18(1):116. https://doi.org/10.1186/s12943-019-1041-z.
Chen W, Zhang B, Chang X. Emerging roles of circular RNAs in osteoporosis. J Cell Mol Med. 2021;25(19):9089–101. https://doi.org/10.1111/jcmm.16906.
Tare RS, Mitchell PD, Kanczler J, Oreffo RO. Isolation, differentiation, and characterisation of skeletal stem cells from human bone marrow in vitro and in vivo. Methods Mol Biol. 2012;816:83–99. https://doi.org/10.1007/978-1-61779-415-5_7.
Chen Y, Yang F, Fang E, Xiao W, Mei H, Li H, Li D, Song H, Wang J, Hong M, Wang X, Huang K, Zheng L, Tong Q. Circular RNA circAGO2 drives cancer progression through facilitating HuR-repressed functions of AGO2-miRNA complexes. Cell Death Differ. 2019;26(7):1346–64. https://doi.org/10.1038/s41418-018-0220-6.
Petkovic S, Muller S. RNA circularization strategies in vivo and in vitro. Nucleic Acids Res. 2015;43(4):2454–65. https://doi.org/10.1093/nar/gkv045.
Li CJ, Cheng P, Liang MK, Chen YS, Lu Q, Wang JY, Xia ZY, Zhou HD, Cao X, Xie H, Liao EY, Luo XH. MicroRNA-188 regulates age-related switch between osteoblast and adipocyte differentiation. J Clin Invest. 2015;125(4):1509–22. https://doi.org/10.1172/JCI77716.
Patop IL, Wust S, Kadener S. Past, present, and future of circRNAs. EMBO J. 2019;38(16): e100836. https://doi.org/10.15252/embj.2018100836.
Kim HS, Wilce MC, Yoga YM, Pendini NR, Gunzburg MJ, Cowieson NP, Wilson GM, Williams BR, Gorospe M, Wilce JA. Different modes of interaction by TIAR and HuR with target RNA and DNA. Nucleic Acids Res. 2011;39(3):1117–30. https://doi.org/10.1093/nar/gkq837.
Lebedeva S, Jens M, Theil K, Schwanhausser B, Selbach M, Landthaler M, Rajewsky N. Transcriptome-wide analysis of regulatory interactions of the RNA-binding protein HuR. Mol Cell. 2011;43(3):340–52. https://doi.org/10.1016/j.molcel.2011.06.008.
Nabors LB, Gillespie GY, Harkins L, King PH. HuR, a RNA stability factor, is expressed in malignant brain tumors and binds to adenine- and uridine-rich elements within the 3’ untranslated regions of cytokine and angiogenic factor mRNAs. Can Res. 2001;61(5):2154–61.
Mazan-Mamczarz K, Galban S, Lopez de Silanes I, Martindale JL, Atasoy U, Keene JD, Gorospe M. RNA-binding protein HuR enhances p53 translation in response to ultraviolet light irradiation. Proc Natl Acad Sci USA. 2003;100(14):8354–9. https://doi.org/10.1073/pnas.1432104100.
Cui Y, Niziolek PJ, MacDonald BT, Zylstra CR, Alenina N, Robinson DR, Zhong Z, Matthes S, Jacobsen CM, Conlon RA, Brommage R, Liu Q, Mseeh F, Powell DR, Yang QM, Zambrowicz B, Gerrits H, Gossen JA, He X, Bader M, Williams BO, Warman ML, Robling AG. Lrp5 functions in bone to regulate bone mass. Nat Med. 2011;17(6):684–91. https://doi.org/10.1038/nm.2388.
Li X, Yang L, Chen LL. The Biogenesis, Functions, and Challenges of Circular RNAs. Mol Cell. 2018;71(3):428–42. https://doi.org/10.1016/j.molcel.2018.06.034.
Huang Y, Xie J, Li E. Comprehensive circular RNA profiling reveals circ_0002060 as a potential diagnostic biomarkers for osteoporosis. J Cell Biochem. 2019;120(9):15688–94. https://doi.org/10.1002/jcb.28838.
Yu L, Liu Y. circRNA_0016624 could sponge miR-98 to regulate BMP2 expression in postmenopausal osteoporosis. Biochem Biophys Res Commun. 2019;516(2):546–50. https://doi.org/10.1016/j.bbrc.2019.06.087.
Huang Y, Xiao D, Huang S, Zhuang J, Zheng X, Chang Y, Yin D. Circular RNA YAP1 attenuates osteoporosis through up-regulation of YAP1 and activation of Wnt/beta-catenin pathway. Biomed Pharmacother. 2020;129:110365. https://doi.org/10.1016/j.biopha.2020.110365.
Chia W, Liu J, Huang YG, Zhang C. A circular RNA derived from DAB1 promotes cell proliferation and osteogenic differentiation of BMSCs via RBPJ/DAB1 axis. Cell Death Dis. 2020;11(5):372. https://doi.org/10.1038/s41419-020-2572-3.
Zhi F, Ding Y, Wang R, Yang Y, Luo K, Hua F. Exosomal hsa_circ_0006859 is a potential biomarker for postmenopausal osteoporosis and enhances adipogenic versus osteogenic differentiation in human bone marrow mesenchymal stem cells by sponging miR-431-5p. Stem Cell Res Ther. 2021;12(1):157. https://doi.org/10.1186/s13287-021-02214-y.
Pereira B, Billaud M, Almeida R. RNA-Binding Proteins in Cancer: Old Players and New Actors. Trends Cancer. 2017;3(7):506–28. https://doi.org/10.1016/j.trecan.2017.05.003.
Zhu YJ, Zheng B, Luo GJ, Ma XK, Lu XY, Lin XM, Yang S, Zhao Q, Wu T, Li ZX, Liu XL, Wu R, Liu JF, Ge Y, Yang L, Wang HY, Chen L. Circular RNAs negatively regulate cancer stem cells by physically binding FMRP against CCAR1 complex in hepatocellular carcinoma. Theranostics. 2019;9(12):3526–40. https://doi.org/10.7150/thno.32796.
Garikipati VNS, Verma SK, Cheng Z, Liang D, Truongcao MM, Cimini M, Yue Y, Huang G, Wang C, Benedict C, Tang Y, Mallaredy V, Ibetti J, Grisanti L, Schumacher SM, Gao E, Rajan S, Wilusz JE, Goukassian D, Houser SR, Koch WJ, Kishore R. Circular RNA CircFndc3b modulates cardiac repair after myocardial infarction via FUS/VEGF-A axis. Nat Commun. 2019;10(1):4317. https://doi.org/10.1038/s41467-019-11777-7.
Grammatikakis I, Abdelmohsen K, Gorospe M. Posttranslational control of HuR function. Wiley Interdiscip Rev RNA. 2017. https://doi.org/10.1002/wrna.1372.
Zhang C, Han X, Yang L, Fu J, Sun C, Huang S, Xiao W, Gao Y, Liang Q, Wang X, Luo F, Lu W, Zhou Y. Circular RNA circPPM1F modulates M1 macrophage activation and pancreatic islet inflammation in type 1 diabetes mellitus. Theranostics. 2020;10(24):10908–24. https://doi.org/10.7150/thno.48264.
Liang Y, Wang H, Chen B, Mao Q, Xia W, Zhang T, Song X, Zhang Z, Xu L, Dong G, Jiang F. circDCUN1D4 suppresses tumor metastasis and glycolysis in lung adenocarcinoma by stabilizing TXNIP expression. Mol Therapy Nucleic acids. 2021;23:355–68. https://doi.org/10.1016/j.omtn.2020.11.012.
Estrada K, Styrkarsdottir U, Evangelou E, Hsu YH, Duncan EL, Ntzani EE, Oei L, Albagha OM, Amin N, Kemp JP, Koller DL, Li G, Liu CT, Minster RL, Moayyeri A, Vandenput L, Willner D, Xiao SM, Yerges-Armstrong LM, Zheng HF, Alonso N, Eriksson J, Kammerer CM, Kaptoge SK, Leo PJ, Thorleifsson G, Wilson SG, Wilson JF, Aalto V, Alen M, Aragaki AK, Aspelund T, Center JR, Dailiana Z, Duggan DJ, Garcia M, Garcia-Giralt N, Giroux S, Hallmans G, Hocking LJ, Husted LB, Jameson KA, Khusainova R, Kim GS, Kooperberg C, Koromila T, Kruk M, Laaksonen M, Lacroix AZ, Lee SH, Leung PC, Lewis JR, Masi L, Mencej-Bedrac S, Nguyen TV, Nogues X, Patel MS, Prezelj J, Rose LM, Scollen S, Siggeirsdottir K, Smith AV, Svensson O, Trompet S, Trummer O, van Schoor NM, Woo J, Zhu K, Balcells S, Brandi ML, Buckley BM, Cheng S, Christiansen C, Cooper C, Dedoussis G, Ford I, Frost M, Goltzman D, Gonzalez-Macias J, Kahonen M, Karlsson M, Khusnutdinova E, Koh JM, Kollia P, Langdahl BL, Leslie WD, Lips P, Ljunggren O, Lorenc RS, Marc J, Mellstrom D, Obermayer-Pietsch B, Olmos JM, Pettersson-Kymmer U, Reid DM, Riancho JA, Ridker PM, Rousseau F, Slagboom PE, Tang NL, Urreizti R, Van Hul W, Viikari J, Zarrabeitia MT, Aulchenko YS, Castano-Betancourt M, Grundberg E, Herrera L, Ingvarsson T, Johannsdottir H, Kwan T, Li R, Luben R, Medina-Gomez C, Palsson ST, Reppe S, Rotter JI, Sigurdsson G, van Meurs JB, Verlaan D, Williams FM, Wood AR, Zhou Y, Gautvik KM, Pastinen T, Raychaudhuri S, Cauley JA, Chasman DI, Clark GR, Cummings SR, Danoy P, Dennison EM, Eastell R, Eisman JA, Gudnason V, Hofman A, Jackson RD, Jones G, Jukema JW, Khaw KT, Lehtimaki T, Liu Y, Lorentzon M, McCloskey E, Mitchell BD, Nandakumar K, Nicholson GC, Oostra BA, Peacock M, Pols HA, Prince RL, Raitakari O, Reid IR, Robbins J, Sambrook PN, Sham PC, Shuldiner AR, Tylavsky FA, van Duijn CM, Wareham NJ, Cupples LA, Econs MJ, Evans DM, Harris TB, Kung AW, Psaty BM, Reeve J, Spector TD, Streeten EA, Zillikens MC, Thorsteinsdottir U, Ohlsson C, Karasik D, Richards JB, Brown MA, Stefansson K, Uitterlinden AG, Ralston SH, Ioannidis JP, Kiel DP, Rivadeneira F. Genome-wide meta-analysis identifies 56 bone mineral density loci and reveals 14 loci associated with risk of fracture. Nat Genet. 2012;44(5):491–501. https://doi.org/10.1038/ng.2249.
Richards JB, Rivadeneira F, Inouye M, Pastinen TM, Soranzo N, Wilson SG, Andrew T, Falchi M, Gwilliam R, Ahmadi KR, Valdes AM, Arp P, Whittaker P, Verlaan DJ, Jhamai M, Kumanduri V, Moorhouse M, van Meurs JB, Hofman A, Pols HA, Hart D, Zhai G, Kato BS, Mullin BH, Zhang F, Deloukas P, Uitterlinden AG, Spector TD. Bone mineral density, osteoporosis, and osteoporotic fractures: a genome-wide association study. Lancet. 2008;371(9623):1505–12. https://doi.org/10.1016/S0140-6736(08)60599-1.
Gong Y, Slee RB, Fukai N, Rawadi G, Roman-Roman S, Reginato AM, Wang H, Cundy T, Glorieux FH, Lev D, Zacharin M, Oexle K, Marcelino J, Suwairi W, Heeger S, Sabatakos G, Apte S, Adkins WN, Allgrove J, Arslan-Kirchner M, Batch JA, Beighton P, Black GC, Boles RG, Boon LM, Borrone C, Brunner HG, Carle GF, Dallapiccola B, De Paepe A, Floege B, Halfhide ML, Hall B, Hennekam RC, Hirose T, Jans A, Juppner H, Kim CA, Keppler-Noreuil K, Kohlschuetter A, LaCombe D, Lambert M, Lemyre E, Letteboer T, Peltonen L, Ramesar RS, Romanengo M, Somer H, Steichen-Gersdorf E, Steinmann B, Sullivan B, Superti-Furga A, Swoboda W, van den Boogaard MJ, Van Hul W, Vikkula M, Votruba M, Zabel B, Garcia T, Baron R, Olsen BR, Warman ML, Osteoporosis-Pseudoglioma Syndrome Collaborative G. LDL receptor-related protein 5 (LRP5) affects bone accrual and eye development. Cell. 2001;107(4):513–23. https://doi.org/10.1016/s0092-8674(01)00571-2.
Lin J, Zheng Z, Liu J, Yang G, Leng L, Wang H, Qiu G, Wu Z. LRP5-Mediated Lipid Uptake Modulates Osteogenic Differentiation of Bone Marrow Mesenchymal Stromal Cells. Front Cell Dev Biol. 2021;9: 766815. https://doi.org/10.3389/fcell.2021.766815.
Wan Y, Lu C, Cao J, Zhou R, Yao Y, Yu J, Zhang L, Zhao H, Li H, Zhao J, Zhu X, He L, Liu Y, Yao Z, Yang X, Guo X. Osteoblastic Wnts differentially regulate bone remodeling and the maintenance of bone marrow mesenchymal stem cells. Bone. 2013;55(1):258–67. https://doi.org/10.1016/j.bone.2012.12.052.
Shen L, Glowacki J, Zhou S. Inhibition of adipocytogenesis by canonical WNT signaling in human mesenchymal stem cells. Exp Cell Res. 2011;317(13):1796–803. https://doi.org/10.1016/j.yexcr.2011.05.018.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
All authors guaranteed the integrity of the entire study. The experiments were conducted by Gengyan Liu, Jia Luo and Zhengguang Wang. Clinical studies were conducted by Yong Li, Gengyan Liu and Jia Luo. Data was analyzed by Zhou Yong. Manuscript was prepared and reviewed by Yong Li. All authors have read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interests.
Ethical standards
The protocol of our study for human were followed with the ethicaal standards of the institutional committee of The Third Xiangya Hospital (Approve no. 2017-R17021) and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. The use of animals in our study was approved and we strictly followed the ethical standards of the ethics committee of The Third Xiangya Hospital.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.

13577_2022_798_MOESM1_ESM.jpg
Supplementary file1 (JPG 1352 KB) Supplementary Figure 1. The effect of circZNF367 overexpression on proliferation, colony formation, apoptosis and bone formation of BMSCs. A, hBMSCs were transduced with circZNF367 expression lentivirus or EV control, then relative cell viability at indicated times was evaluated by CCK-8 assay. B–C, hBMSCs (4000/well) transduced with circZNF367 expression lentivirus or EV control were used for colony formation assay (B). Number of colony number per well was shown (C). D–E, hBMSCs were transduced with circZNF367 expression lentivirus or EV control, then stained with Annexin-V FITC and PI for flow cytometry analysis (D). The percentage of Annexin-V apoptotic cells was shown (E). All assays were done in triplicates. *P< 0.05

13577_2022_798_MOESM2_ESM.jpg
Supplementary file2 (JPG 664 KB) Supplementary Figure 2. The influence of circZNF367 overexpression on adipogenic differentiation markers of BMSCs. hBMSCs transduced with circZNF367 expression lentivirus or EV control were cultured with adipogenic-inducing medium for 14 days, then relative CEBPα and PPARγ expression were evaluated by qRT-PCR (H) and western blot (I). n.s. = not significant

13577_2022_798_MOESM3_ESM.jpg
Supplementary file3 (JPG 1573 KB) Supplementary Figure 3. The effect of circZNF367 knockdown on proliferation, colony formation, apoptosis and bone formation of BMSCs. A, hBMSCs were transduced with sh-circ-1, sh-circ-2 or sh-NC lentivirus vector, then relative cell viability at indicated times was evaluated by CCK-8 assay. B–C, hBMSCs (4000/well) transduced with sh-circ-1, sh-circ-2 or sh-NC lentivirus vector were used for colony formation assay (B). Number of colony number per well was shown (C). D–E, hBMSCs were transduced with sh-circ-1, sh-circ-2 or sh-NC lentivirus vector, then stained with Annexin-V FITC and PI for flow cytometry analysis (D). The percentage of Annexin-V apoptotic cells was shown (E). All assays were done in triplicates. *P< 0.05

13577_2022_798_MOESM4_ESM.jpg
Supplementary file4 (JPG 1352 KB) Supplementary Figure S4. The influence of cricZNF367 on the downstream targets of Wnt/β-catenin signaling. A–B, Gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of hBMSCs transduced with circZNF367 expression lentivirus or EV control. C, hBMSCs were transduced with circZNF367 expression lentivirus or EV control, then relative LRP5 expression was evaluated by qRT-PCR. D–E, hBMSCs were transduced with circZNF367, EV, sh-circ-1, sh-circ-2 or sh-NC vector, then relative expression of indicated genes were evaluated by qRT-PCR. All assays were done in triplicates. *P< 0.05
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liu, G., Luo, J., Wang, Z. et al. CircZNF367 suppresses osteogenic differentiation of human bone marrow mesenchymal stromal/stem cells via reducing HuR-mediated mRNA stability of LRP5. Human Cell 36, 146–162 (2023). https://doi.org/10.1007/s13577-022-00798-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13577-022-00798-y