Abstract
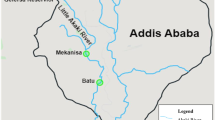
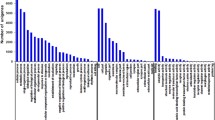
The Korean Demilitarized Zone (DMZ) has almost completely protected surface water conditions. Despite the water conditions of DMZ are excellent clear, downstream of river or natural irrigation ponds near the region are recently developed by human activities such as agriculture, ginseng fields or ecotourism. To understand the potential effect of physico-chemical characteristics of the DMZ water, the freshwater model Daphnia magna was exposed to water samples that were collected from four sites near the DMZ. RNA-seq identified some differentially expressed transcripts which are sensitive to small differences of water chemistry. Particularly transcripts coding for the stress responsive heat shock protein (Hsp) superfamily and innate immune system were significantly increased in response to the water samples of irrigation ponds and downstream of Seweol river where are analyzed to have low pH and high levels of chemical fertilizer nutrients such as nitrogen (N) and phosphorus (P). These water samples decreased mRNA expressions of crucial members which are involved in crustacean morphogenesis and reproduction metabolism (e.g. cuticle protein family, ecdysteroid metabolism, vitellogenesis). Evidence on the high transcriptional sensitivity of the aquatic model D. magna against small differences of water chemistry will be a useful strength for further applications in environmental and ecotoxicological genomics.
Similar content being viewed by others
References
John, K. H. The Korean DMZ: a fragile ecosystem. Science 280, 808–809 (1998).
Healy, H. Korean demilitarized zone: Peace and nature park. Int. J. World Peace 24, 61–83 (2007).
Smith, V. H., Tilman, G. D. & Nekola, J. C. Eutrophication: impacts of excess nutrient inputs on freshwater, marine, and terrestrial ecosystems. Environ. Pollut. 100, 179–196 (1999).
Correll, D. L. The role of phosphorus in the eutrophication receiving waters: a review. J. Environ. Qual. 27, 261–266 (1998).
Baird, D. J., Barber, I., Bradley, M., Calow, P. & Soares, A. M. V. M. The Daphnia bioassay: a critique. Hydrobiologia 188-189, 403–406 (1989).
Shaw, J. R. et al. in Recent Advances in Experimental Biology, Toxicogenomics, vol. 2 (edn Hogstrand, C. & Kille, P.) 165–219 (Elsevier, London, U.K, 2008).
Frydman, J. Folding of newly translated proteins in vivo: the role of molecular chaperones. Annu. Rev. Biochem. 70, 603–649 (2001).
Hartl, F. U. & Hayer-Hartl, M. Molecular chaperones in the cytosol: from nascent chain to folded protein. Science 295, 1852–1858 (2002).
Feder, M. E. & Hofmann, G. E. Heat shock proteins, molecular chaperones, and the stress response: evolutionary and ecological physiology. Annu. Rev. Physiol. 61, 243–282 (1999).
Akira, S., Uematsu, S. & Takeuchi, O. Pathogen recognition and innate immunity. Cell 124, 783–801 (2006).
Iwanaga, S. & Lee, B. L. Recent advances in the innate immunity of invertebrate animals. J. Biochem. Mol. Biol. 38, 128–150 (2005).
McTaggart, S. J., Conlon, C., Colbourne, J. K., Blaxter, M. L. & Little, T. J. The components of the Daphnia pulex immune system as revealed by complete genome sequencing. BMC Genomics 10, 1 (2009).
Decaestecker, E., Labbé, P., Ellegaard, K., Allen, J. E. & Little, T. J. Candidate innate immune system gene expression in the ecological model Daphnia. Dev. Comp. Immunol. 35, 1068–1077 (2011).
Bang, S. H. et al. Acute and chronic toxicity assessment and the gene expression of Dhb, Vtg, Arnt, CYP4, and CYP314 in Daphnia magna exposed to pharmaceuticals. Mol. Cell. Toxicol. 11, 153–160 (2015).
Tokishita, S. et al. Organization and repression by juvenile hormone of a vitellogenin gene cluster in the crustacean, Daphnia magna. Biochem. Biophys. Res. Commun. 345, 362–370 (2006).
Hannas, B. R. et al. Regulation and dysregulation of vitellogenin mRNA accumulation in daphnids (Daphnia magna). Aquat. Toxicol. 101, 351–357 (2011).
Martin-Creuzburg, D., Westerlund, S. A. & Hoffmann, K. H. Ecdysteroid levels in Daphnia magna during a molt cycle: determination by radioimmunoassay (RIA) and liquid chromatography-mass spectrometry (LC-MS). Gen. Comp. Endocrinol. 151, 66–71 (2007).
Kim, Y.-J., Lee, N., Woo, S., Ryu, J.-C. & Yum, S. Transcriptomic change as evidence for cadmium-induced endocrine disruption in marine fish model of medaka, Oryzias javanicus. Mol. Cell. Toxicol. 12, 409–420 (2016).
Grabherr, M. G. et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652 (2011).
Pertea, G. et al. TIGR Gene Indices clustering tools (TGICL): a software system for fast clustering of large EST datasets. Bioinformatics 19, 651–652 (2003).
Conesa, A. et al. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676 (2005).
Haas, B. J. et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494–1512 (2013).
Schultz, J., Copley, R. R., Doerks, T., Ponting, C. P. & Bork, P. SMART: a web-based tool for the study of genetically mobile domains. Nucleic Acids Res. 28, 231–234 (2000).
Thomas, P. D. et al. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 13, 2129–2141 (2003).
Bateman, A. et al. The Pfam protein families database. Nucleic Acids Res. 32, 138–141 (2004).
Bru, C. et al. The ProDom database of protein domain families: more emphasis on 3D. Nucleic Acids Res. 33, 212–215 (2005).
Yeats, C. et al. Gene3D: modelling protein structure, function and evolution. Nucleic Acids Res. 34, 281–284 (2006).
Li, B. & Dewey, C. N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Sun, J., Nishiyama, T., Shimizu, K. & Kadota, K. TCC: an R package for comparing tag count data with robust normalization strategies. BMC Bioinformatics 14, 219 (2013).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kim, BM., Ahn, S.H., Choi, N.R. et al. Transcriptome profiles of Daphnia magna across to the different water chemistry of surface water of the Korean Demilitarized Zone. Toxicol. Environ. Health Sci. 9, 188–198 (2017). https://doi.org/10.1007/s13530-017-0320-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13530-017-0320-6