Abstract
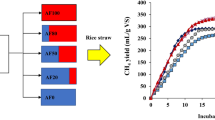
Anaerobic lignocellulosic microbial consortia are known to be prodigiously efficient at converting lignocellulosic biomass to methane. In this study, the efficacy of anaerobic fungal consortia (AFC) from five different inocula, including Bubalus bubalis rumen fluid (RU), in degrading filter paper, microcrystalline cellulose, and rice straw (RS), was screened. The AFC from RU performed best in lignocellulosic material degradation and methane production; thus, RU was selected for further experiments. Consecutive batch subculturing (CBS) was performed in RU to enrich and stabilize the dominant and key microorganisms categorized as anaerobic fungi, using the addition of antibacterial agents to suppress the growth of untargeted bacteria. After the CBS, subculture E19 proved the most efficient, with RS degradation of 84% and a methane yield of 310 mL/g VSadded, representing 1.83- and 2.25-fold increases compared to the initial seed, respectively. The microbial community of E19 consisted of anaerobic fungi (uncultured Neocallimastigales, Anaeromyces sp., Orpinomyces sp., and Feramyces sp.) coexisting with anaerobic bacteria (streptomycin resistant Proteiniphilum acetatigenes), and methanogens. The E19 consortium was able to use various carbon sources (87.5%) and contained potential genes encoding enzymes involved in RS degradation. The microbial community of E19 was highly stable, making it a promising inoculum for biomass degradation, especially for anaerobic digestion to produce biogas.





Similar content being viewed by others
Abbreviations
- 16S rRNA:
-
16S ribosomal RNA is the RNA component of the 30S subunit of a prokaryotic ribosome
- AB:
-
Anaerobic bacteria
- AD:
-
Anaerobic digestion
- AF:
-
Anaerobic fungi
- AFC:
-
Efficacy of anaerobic fungal consortia
- ALMC:
-
Anaerobic lignocellulosic microbial consortium
- AMT:
-
Acetoclastic methanogens
- ANOVA:
-
One-way analysis of variance
- AOAC:
-
Association of Official Agricultural Chemists
- APHA:
-
American Public Health Association
- B-ARISA:
-
Automated method of ribosomal intergenic spacer analysis for bacteria
- BMP:
-
Biochemical methane potential
- BUSCO:
-
Benchmarking Universal Single-Copy Orthologs
- CAZy:
-
Carbohydrate-active enzymes
- CBS:
-
Consecutive batch subculturing
- CM:
-
Cow manure
- COD:
-
Chemical oxygen demand
- CSTR:
-
Continuously stirred tank reactor
- E1 − E19:
-
Serial number of subculture during enrichment and stabilization
- F-ARISA:
-
Automated method of ribosomal intergenic spacer analysis for fungi
- FP:
-
Filter paper
- GC:
-
Gas chromatography
- GH:
-
Glycoside hydrolase
- GM:
-
Goat manure
- HMT:
-
Hydrogenotrophic methanogens
- ISR:
-
The inoculum (I) to substrate (S) ratio
- ITS:
-
Internal transcribed spacer
- MCC:
-
Microcrystalline cellulose
- MT:
-
Methanigens
- MS:
-
Microbial sludge from anaerobic wastewater treatment system of a palm oil mill factory
- PCR:
-
Polymerase chain reaction
- PM:
-
Pig manure
- qPCR:
-
Quantitative real-time PCR
- RS:
-
Rice straw
- RU:
-
Rumen fluid
- SSU:
-
Small subunit
- TS:
-
Total solids, defined as mass remaining after drying at 105 °C
- VFAs:
-
Volatile fatty acids
- VS:
-
Volatile solids, determined as weight loss from heating in air at 550 °C
- °C:
-
Degree celsius (temperature unit)
- bp:
-
Base pairs (nucleic acid unit)
- g :
-
Gram (mass unit)
- h :
-
Hour (time unit)
- L :
-
Liter (volume unit)
- m :
-
Meter (length unit)
- μm:
-
Micrometer (length unit)
- mL:
-
Milliliter (volume unit)
- mg:
-
Milligram (mass unit)
- mm:
-
Millimeter (length unit)
- mM:
-
Millimolar (concentration unit)
- min:
-
Minute (time unit)
- M:
-
Molar (concentration unit)
- s:
-
Second (time unit)
References
Tursi A (2019) A review on biomass: importance, chemistry, classification, and conversion. Biofuel Res J 6(2):962–979. https://doi.org/10.18331/brj2019.6.2.3
Theuerl S, Klang J, Prochnow A (2019) Process disturbances in agricultural biogas production-causes, mechanisms, and effects on the biogas microbiome: a review. Energies 12(365):20. https://doi.org/10.3390/en12030365
Office of Agricultural Economics OAE (2020) Information of agricultural production in Thailand http://www.oae.go.th/view/1/Information/EN-US. Accessed 25 Jun 2019
Monlau F, Barakat A, Trably E, Dumas C, Steyer JP, Carrère H (2013) Lignocellulosic materials into biohydrogen and biomethane: impact of structural features and pretreatment. Crit Rev Environ Sci Technol 43(3):260–322. https://doi.org/10.1080/10643389.2011.604258
Sun C, Liu R, Cao W, Yin R, Mei Y, Zhang L (2015) Impacts of alkaline hydrogen peroxide pretreatment on chemical composition and biochemical methane potential of agricultural crop stalks. Energy Fuels 29(8):4966–4975. https://doi.org/10.1021/acs.energyfuels.5b00838
Widjaja T, Noviyanto AA, Gunawan S (2016) The effect of rumen and mixed microorganism (rumen and effective microorganisms) on biogas production from rice straw waste. ARPN JEAS 11(4):2702–2710
Contreras LM, Schelle H, Sebrango CR, Pereda I (2012) Methane potential and biodegradability of rice straw, rice husk and rice residues from the drying process. Water Sci Technol 65(6):1142–1149. https://doi.org/10.2166/wst.2012.951
Dehghani M, Karimi K, Sadeghi M (2015) Pretreatment of rice straw for the improvement of biogas production. Energy Fuels 29(6):3770–3775. https://doi.org/10.1021/acs.energyfuels.5b00718
Mustafa AM, Poulsen TG, Xia Y, Sheng K (2017) Combinations of fungal and milling pretreatments for enhancing rice straw biogas production during solid-state anaerobic digestion. Biores Technol 224:174–182. https://doi.org/10.1016/j.biortech.2016.11.028
Doi RH (2008) Cellulases of mesophilic microorganisms: cellulosome and noncellulosome producers. Ann N Y Acad Sci 1125:267–279. https://doi.org/10.1196/annals.1419.002
Dollhofer V, Podmirseg SM, Callaghan TM, Griffith GW, Fliegerova K (2015) Anaerobic fungi and their potential for biogas production. Adv Biochem Eng Biotechnol 151:41–61. https://doi.org/10.1007/978-3-319-21993-6_2
Youssef N, Couger M, Struchtemeyer C, Liggenstoffer A, Prade R, Najar F, Atiyeh H, Wilkins M, Elshahed M (2013) The genome of the anaerobic fungus Orpinomyces sp. strain C1A reveals the unique evolutionary history of a remarkable plant biomass degrader. Appl Environ Microbiol 79(15):4620–4634. https://doi.org/10.1128/AEM.00821-13
Tabatabaei M, Aghbashlo M, Valijanian E, Kazemi Shariat Panahi H, Nizami A-S, Ghanavati H, Sulaiman A, Mirmohamadsadeghi S, Karimi K (2020) A comprehensive review on recent biological innovations to improve biogas production, Part 1: upstream strategies. Renew Energy 146:1204–1220. https://doi.org/10.1016/j.renene.2019.07.037
Tabatabaei M, Aghbashlo M, Valijanian E, Kazemi Shariat Panahi H, Nizami A-S, Ghanavati H, Sulaiman A, Mirmohamadsadeghi S, Karimi K (2020) A comprehensive review on recent biological innovations to improve biogas production, Part 2: mainstream and downstream strategies. Renew Energy 146:1392–1407. https://doi.org/10.1016/j.renene.2019.07.047
Thongbunrod N, Chaiprasert P (2021) Efficacy and metagenomic analysis of the stabilized anaerobic lignocellulolytic microbial consortium from Bubalus bubalis rumen with rice straw enrichment for methane production. BioEnergy Research 14(3):870–890. https://doi.org/10.1007/s12155-020-10167-y
Rouches E, Zhou S, Steyer JP, Carrere H (2016) White-rot fungi pretreatment of lignocellulosic biomass for anaerobic digestion: impact of glucose supplementation. Process Biochem 51(11):1784–1792. https://doi.org/10.1016/j.procbio.2016.02.003
Kainthola J, Kalamdhad AS, Goud VV, Goel R (2019) Fungal pretreatment and associated kinetics of rice straw hydrolysis to accelerate methane yield from anaerobic digestion. Bioresour Technol 286:121368. https://doi.org/10.1016/j.biortech.2019.121368
Kong X, Du J, Ye X, Xi Y, Jin H, Zhang M, Guo D (2018) Enhanced methane production from wheat straw with the assistance of lignocellulolytic microbial consortium TC-5. Bioresour Technol 263:33–39. https://doi.org/10.1016/j.biortech.2018.04.079
Li P, He C, Li G, Ding P, Lan M, Gao Z, Jiao Y (2020) Biological pretreatment of corn straw for enhancing degradation efficiency and biogas production. Bioengineered 11(1):251–260. https://doi.org/10.1080/21655979.2020.1733733
Xu W, Fu S, Yang Z, Lu J, Guo R (2018) Improved methane production from corn straw by microaerobic pretreatment with a pure bacteria system. Bioresour Technol 259:18–23. https://doi.org/10.1016/j.biortech.2018.02.046
Li J, Wu Y, Zhao J, Wang S, Dong Z, Shao T (2022) Bioaugmented degradation of rice straw combining two novel microbial consortia and lactic acid bacteria for enhancing the methane production. Bioresour Technol 344:126148. https://doi.org/10.1016/j.biortech.2021.126148
Ozbayram EG, Akyol C, Ince B, Karakoc C, Ince O (2018) Rumen bacteria at work: bioaugmentation strategies to enhance biogas production from cow manure. J Appl Microbiol 124(2):491–502. https://doi.org/10.1111/jam.13668
Ozbayram EG, Kleinsteuber S, Nikolausz M, Ince B, Ince O (2017) Effect of bioaugmentation by cellulolytic bacteria enriched from sheep rumen on methane production from wheat straw. Anaerobe 46:122–130. https://doi.org/10.1016/j.anaerobe.2017.03.013
Ma Y, Li Y, Li Y, Cheng Y, Zhu W (2020) The enrichment of anaerobic fungi and methanogens showed higher lignocellulose degrading and methane producing ability than that of bacteria and methanogens. World J Microbiol Biotechnol 36(125):1–9. https://doi.org/10.1007/s11274-020-02894-3
Tsapekos P, Kougias PG, Vasileiou SA, Treu L, Campanaro S, Lyberatos G, Angelidaki I (2017) Bioaugmentation with hydrolytic microbes to improve the anaerobic biodegradability of lignocellulosic agricultural residues. Bioresour Technol 234:350–359. https://doi.org/10.1016/j.biortech.2017.03.043
Yıldırım E, Ince O, Aydin S, Ince B (2017) Improvement of biogas potential of anaerobic digesters using rumen fungi. Renew Energy 109:346–353. https://doi.org/10.1016/j.renene.2017.03.021
Akyol C, Ince O, Bozan M, Ozbayram EG, Ince B (2019) Fungal bioaugmentation of anaerobic digesters fed with lignocellulosic biomass: what to expect from anaerobic fungus Orpinomyces sp. Bioresour Technol 277:1–10. https://doi.org/10.1016/j.biortech.2019.01.024
Ferraro A, Dottorini G, Massini G, Mazzurco Miritana V, Signorini A, Lembo G, Fabbricino M (2018) Combined bioaugmentation with anaerobic ruminal fungi and fermentative bacteria to enhance biogas production from wheat straw and mushroom spent straw. Bioresour Technol 260:364–373. https://doi.org/10.1016/j.biortech.2018.03.128
Wei YQ, Long RJ, Yang H, Yang HJ, Shen XH, Shi RF, Wang ZY, Du JG, Qi XJ, Ye QH (2016) Fiber degradation potential of natural co-cultures of Neocallimastix frontalis and Methanobrevibacter ruminantium isolated from yaks (Bos grunniens) grazing on the Qinghai Tibetan Plateau. Anaerobe 39:158–164. https://doi.org/10.1016/j.anaerobe.2016.03.005
Nguyen QH, Le PD, Chim C, Le ND, Fievez V (2019) Potential to mitigate ammonia emission from slurry by increasing dietary fermentable fiber through inclusion of tropical byproducts in practical diets for growing pigs. Asian-Australas J Anim Sci 32(4):574–584. https://doi.org/10.5713/ajas.18.0481
Grenet E, Bernalier A, Jamot J, Fonty G (1993) Degradation of untreated and anhydrous ammonia-treated wheat straw by two strains of rumen anaerobic fungi. Ann Zootech 42(180):1–1
Ha JK, Lee SS, Kim SW, Han IK, Ushida K, Cheng KJ (2001) Degradation of rice straw by rumen fungi and cellulolytic bacteria through mono-, co- or sequential- culture. Asian Australas J Anim Sci 14(6):797–802
APHA (2012) Standard methods for the examination of water and wastewater, 22nd edition out of print. American public health association (APHA), American water works association (AWWA) and water environment federation (WEF), Washington
Bauchop T (1979) Rumen anaerobic fungi of cattle and sheep. Appl Environ Microbiol 38(1):148–158
Mertens DR (2002) Gravimetric determination of amylase-treated neutral detergent fiber in feeds with refluxing in beakers or crucibles: collaborative study. J AOAC Int 85:24
Theodorou M, Williams B, Dhanoa M, McAllan A, France J (1994) A simple gas production method using a pressure transducer to determine the fermentation kinetics of ruminant feeds. Journal of Animal and Feed Sciences 48(3):185–197. https://doi.org/10.1016/0377-8401(94)90171-6
Panichnumsin P, Nopharatana A, Ahring B, Chaiprasert P (2010) Production of methane by co-digestion of cassava pulp with various concentrations of pig manure. Biomass Bioenergy 34(8):1117–1124. https://doi.org/10.1016/j.biombioe.2010.02.018
Ranjard L, Poly F, Lata JC, Mougel C, Thioulouse J, Nazaret S (2001) Characterization of bacterial and fungal soil communities by automated ribosomal intergenic spacer analysis fingerprints: biological and methodological variability. Appl Environ Microbiol 67(10):4479–4487. https://doi.org/10.1128/aem.67.10.4479-4487.2001
Kittelmann S, Naylor GE, Koolaard JP, Janssen PH (2012) A proposed taxonomy of anaerobic fungi (class neocallimastigomycetes) suitable for large-scale sequence-based community structure analysis. PLoS ONE 7(5):1–3. https://doi.org/10.1371/journal.pone.0036866
Yu Q, Liu R, Li K, Ma R (2019) A review of crop straw pretreatment methods for biogas production by anaerobic digestion in China. Renew Sustain Energy Rev 107:51–58. https://doi.org/10.1016/j.rser.2019.02.020
Belda-Galbis CM, Pina-Perez MC, Espinosa J, Marco-Celdran A, Martinez A, Rodrigo D (2014) Use of the modified Gompertz equation to assess the Stevia rebaudiana Bertoni antilisterial kinetics. Food Microbiol 38:56–61. https://doi.org/10.1016/j.fm.2013.08.009
Li Y, Zhang R, Liu G, Chen C, He Y, Liu X (2013) Comparison of methane production potential, biodegradability, and kinetics of different organic substrates. Bioresour Technol 149:565–569. https://doi.org/10.1016/j.biortech.2013.09.063
Baetge S, Kaltschmitt M (2018) Rice straw and rice husks as energy sources-comparison of direct combustion and biogas production. Biomass Conversion and Biorefinery 8(3):719–737. https://doi.org/10.1007/s13399-018-0321-y
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75(23):7537–7541. https://doi.org/10.1128/AEM.01541-09
Silva reference files: Release 132 (2018). https://mothur.org/wiki/silva_reference_files/. Accessed 28 Sept 2020
Kessy A, Allan Z, Timo P, Raivo P, Filipp I, Henrik NR, Urmas K (2020) UNITE mothur release for Fungi 2. UNITE Community. https://unite.ut.ee/repository.php. Accessed 25 Sep 2020
Koetschan C, Kittelmann S, Lu J, Al-Halbouni D, Jarvis GN, Muller T, Wolf M, Janssen PH (2014) Internal transcribed spacer 1 secondary structure analysis reveals a common core throughout the anaerobic fungi (Neocallimastigomycota). PLoS ONE 9(3):1–10. https://doi.org/10.1371/journal.pone.0091928
Batut B, Hiltemann S, Bagnacani A, Baker D, Bhardwaj V, Blank C, Bretaudeau A, Brillet-Gueguen L, Cech M, Chilton J, Clements D, Doppelt-Azeroual O, Erxleben A, Freeberg MA, Gladman S, Hoogstrate Y, Hotz HR, Houwaart T, Jagtap P, Lariviere D, Le Corguille G, Manke T, Mareuil F, Ramirez F, Ryan D, Sigloch FC, Soranzo N, Wolff J, Videm P, Wolfien M, Wubuli A, Yusuf D, Galaxy Training N, Taylor J, Backofen R, Nekrutenko A, Gruning B (2018) Community-driven data analysis training for biology. Cell Syst 6(6):752–758. https://doi.org/10.1016/j.cels.2018.05.012
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Phan S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA (2012) SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19(5):455–477. https://doi.org/10.1089/cmb.2012.0021
Abubucker S, Segata N, Goll J, Schubert AM, Izard J, Cantarel BL, Rodriguez-Mueller B, Zucker J, Thiagarajan M, Henrissat B, White O, KelleyMethe´SchlossGeversMitrevaHuttenhower TSBPDDMC (2012) Metabolic reconstruction for metagenomic data and its application to the human microbiome. Appl Environ Microbiol 8(6):1–18. https://doi.org/10.1371/journal.pcbi.1002358
Levy Karin E, Mirdita M, Soding J (2020) MetaEuk-sensitive, high-throughput gene discovery, and annotation for large-scale eukaryotic metagenomics. Microbiome 8(48):1–15. https://doi.org/10.1186/s40168-020-00808-x
Keller O, Kollmar M, Stanke M, Waack S (2011) A novel hybrid gene prediction method employing protein multiple sequence alignments. Bioinformatics 27(6):757–763. https://doi.org/10.1093/bioinformatics/btr010
Eko H, Chaiprasert P (2019) Enhancement of methane production from high solid anaerobic digestion of pretreated palm oil decanter cake using a modified solid inclined reactor. J Chem Technol Biotechnol 95(3):781–790. https://doi.org/10.1002/jctb.6266
Liu Y, Whitman WB (2008) Metabolic, phylogenetic, and ecological diversity of the methanogenic archaea. Ann N Y Acad Sci 1125:171–189. https://doi.org/10.1196/annals.1419.019
Ozbayram EG, Kleinsteuber S, Nikolausz M (2020) Biotechnological utilization of animal gut microbiota for valorization of lignocellulosic biomass. Appl Microbiol Biotechnol 104(2):489–508. https://doi.org/10.1007/s00253-019-10239-w
Li K, Zhu H, Zhang Y, Zhang H (2017) Characterization of the microbial communities in rumen fluid inoculated reactors for the biogas digestion of wheat straw. Sustainability 9:243. https://doi.org/10.3390/su9020243
Nagler M, Kozjek K, Etemadi M, Insam H, Podmirseg SM (2019) Simple yet effective: microbial and biotechnological benefits of rumen liquid addition to lignocellulose-degrading biogas plants. J Biotechnol 300:1–10. https://doi.org/10.1016/j.jbiotec.2019.05.004
Joblin KN (1981) Isolation, enumeration, and maintenance of rumen anaerobic fungi in roll tubes. Appl Environ Microbiol 42(6):1119–1122
Jin W, Cheng YF, Mao SY, Zhu WY (2011) Isolation of natural cultures of anaerobic fungi and indigenously associated methanogens from herbivores and their bioconversion of lignocellulosic materials to methane. Bioresour Technol 102(17):7925–7931. https://doi.org/10.1016/j.biortech.2011.06.026
Callaghan TM, Podmirseg SM, Hohlweck D, Edwards JE, Puniya AK, Dagar SS, Griffith GW (2015) Buwchfawromyces eastonii gen. nov., sp. nov.: a new anaerobic fungus (Neocallimastigomycota) isolated from buffalo faeces. MycoKeys 9:11–28. https://doi.org/10.3897/mycokeys.9.9032
Dagar SS, Kumar S, Griffith GW, Edwards JE, Callaghan TM, Singh R, Nagpal AK, Puniya AK (2015) A new anaerobic fungus (Oontomyces anksri gen. nov., sp. nov.) from the digestive tract of the Indian camel (Camelus dromedarius). Fungal Biol Rev 119(8):731–737. https://doi.org/10.1016/j.funbio.2015.04.005
Joshi A, Lanjekar VB, Dhakephalkar PK, Callaghan TM, Griffith GW, Dagar SS (2018) Liebetanzomyces polymorphus gen. et sp. nov., a new anaerobic fungus (Neocallimastigomycota) isolated from the rumen of a goat. MycoKeys 40:89–110. https://doi.org/10.3897/mycokeys.40.28337
Jing R, Yan Y (2020) Metagenomic analysis reveals antibiotic resistance genes in the bovine rumen. Microb Pathog 149:1–25. https://doi.org/10.1016/j.micpath.2020.104350
Treu L, Campanaro S, Kougias PG, Sartori C, Bassani I, Angelidaki I (2018) Hydrogen-fueled microbial pathways in biogas upgrading systems revealed by genome-centric metagenomics. Front Microbiol 9:1–16. https://doi.org/10.3389/fmicb.2018.01079
Ward AJ, Hobbs PJ, Holliman PJ, Jones DL (2008) Optimisation of the anaerobic digestion of agricultural resources. Bioresour Technol 99(17):7928–7940. https://doi.org/10.1016/j.biortech.2008.02.044
Chandra R, Vijay VK, Subbarao PMV, Khura TK (2012) Production of methane from anaerobic digestion of jatropha and pongamia oil cakes. Appl Energy 93:148–159. https://doi.org/10.1016/j.apenergy.2010.10.049
Kim M, Gomec CY, Ahn Y, Speece RE (2003) Hydrolysis and acidogenesis of particulate organic material in mesophilic and thermophilic anaerobic digestion. Environ Technol 24(9):1183–1190. https://doi.org/10.1080/09593330309385659
Siegert I, Banks C (2005) The effect of volatile fatty acid additions on the anaerobic digestion of cellulose and glucose in batch reactors. Process Biochem 40(11):3412–3418. https://doi.org/10.1016/j.procbio.2005.01.025
Theodorou MK, Gill M, King-Spooner C, Beever DE (1990) Enumeration of anaerobic chytridiomycetes as thallus-forming units: novel method for quantification of fibrolytic fungal populations from the digestive tract ecosystem. Appl Environ Microbiol 56(4):1073–1078. https://doi.org/10.1128/aem.56.4.1073-1078.1990
Orpin CG (1977) The rumen flagellate Piromonas communis: its life-history and invasion of plant material in the rumen. J Gen Microbiol 99:107–117. https://doi.org/10.1099/00221287-99-1-107
Hansen JC, Aanderud ZT, Reid LE, Bateman C, Hansen CL, Rogers LS, Hansen LD (2021) Enhancing waste degradation and biogas production by pre-digestion with a hyperthermophilic anaerobic bacterium. Biofuel Res J 8(3):1433–1443. https://doi.org/10.18331/brj2021.8.3.3
Mirmohamadsadeghi S, Karimi K, Azarbaijani R, Parsa Yeganeh L, Angelidaki I, Nizami A-S, Bhat R, Dashora K, Vijay VK, Aghbashlo M, Gupta VK, Tabatabaei M (2021) Pretreatment of lignocelluloses for enhanced biogas production: a review on influencing mechanisms and the importance of microbial diversity. Renew Sustain Energy Rev 135:1–18. https://doi.org/10.1016/j.rser.2020.110173
Griessmeier V, Gescher J (2018) Influence of the potential carbon sources for field denitrification beds on their microbial diversity and the fate of carbon and nitrate. Front Microbiol 9:1–15. https://doi.org/10.3389/fmicb.2018.01313
O’Shea R, Lin R, Wall DM, Browne JD, Murphy JD (2021) Distillery decarbonization and anaerobic digestion: balancing benefits and drawbacks using a compromise programming approach. Biofuel Res J 8(3):1417–1432. https://doi.org/10.18331/brj2021.8.3.2
Garcia SL, Jangid K, Whitman WB, Das KC (2011) Transition of microbial communities during the adaption to anaerobic digestion of carrot waste. Bioresour Technol 102(15):7249–7256. https://doi.org/10.1016/j.biortech.2011.04.098
Ito T, Yoshiguchi K, Ariesyady HD, Okabe S (2011) Identification of a novel acetate-utilizing bacterium belonging to Synergistes group 4 in anaerobic digester sludge. ISME J 5(12):1844–1856. https://doi.org/10.1038/ismej.2011.59
Downes J, Vartoukian SR, Dewhirst FE, Izard J, Chen T, Yu WH, Sutcliffe IC, Wade WG (2009) Pyramidobacter piscolens gen. nov., sp. nov., a member of the phylum ‘Synergistetes’ isolated from the human oral cavity. Int J Syst Evol Microbiol 59(5):972–980. https://doi.org/10.1099/ijs.0.000364-0
Chen S, Dong X (2005) Proteiniphilum acetatigenes gen. nov., sp. nov., from a UASB reactor treating brewery wastewater. Int J Syst Evol Microbiol 55(6):2257–2261. https://doi.org/10.1099/ijs.0.63807-0
Kumar M, Verma S, Gazara RK, Kumar M, Pandey A, Verma PK, Thakur IS (2018) Genomic and proteomic analysis of lignin degrading and polyhydroxyalkanoate accumulating beta-proteobacterium Pandoraea sp. ISTKB Biotechnol Biofuels 11:154. https://doi.org/10.1186/s13068-018-1148-2
Liu D (2015) Aeromonas. In: Mol. Microbiol. pp 1099–1110.https://doi.org/10.1016/b978-0-12-397169-2.00061-5
Wilhelm RC, Singh R, Eltis LD, Mohn WW (2019) Bacterial contributions to delignification and lignocellulose degradation in forest soils with metagenomic and quantitative stable isotope probing. ISME J 13(2):413–429. https://doi.org/10.1038/s41396-018-0279-6
Gruninger RJ, Puniya AK, Callaghan TM, Edwards JE, Youssef N, Dagar SS, Fliegerova K, Griffith GW, Forster R, Tsang A, McAllister T, Elshahed MS (2014) Anaerobic fungi (phylum Neocallimastigomycota): advances in understanding their taxonomy, life cycle, ecology, role, and biotechnological potential. FEMS Microbiol Ecol 90(1):1–17. https://doi.org/10.1111/1574-6941.12383
Hanafy RA, Elshahed MS, Youssef NH (2018) Feramyces austinii, gen. nov., sp. nov., an anaerobic gut fungus from rumen and fecal samples of wild Barbary sheep and fallow deer. Mycologia 110(3):513–525. https://doi.org/10.1080/00275514.2018.1466610
Hanafy RA, Lanjekar VB, Dhakephalkar PK, Callaghan TM, Dagar SS, Griffith GW, Elshahed MS, Youssef NH (2020) Seven new Neocallimastigomycota genera from wild, zoo-housed, and domesticated herbivores greatly expand the taxonomic diversity of the phylum. Mycologia:1–28 https://doi.org/10.1080/00275514.2019.1696619
Kameshwar AKS, Qin W (2018) Genome wide analysis reveals the extrinsic cellulolytic and biohydrogen generating abilities of Neocallimastigomycota fungi. J Genomics 6:74–87. https://doi.org/10.7150/jgen.25648
Peng X, Wilken SE, Lankiewicz TS, Gilmore SP, Brown JL, Henske JK, Swift CL, Salamov A, Barry K, Grigoriev IV, Theodorou MK, Valentine DL, O’Malley MA (2021) Genomic and functional analyses of fungal and bacterial consortia that enable lignocellulose breakdown in goat gut microbiomes. Nat Microbiol 6(4):499–511. https://doi.org/10.1038/s41564-020-00861-0
Hanafy RA, Johnson B, Youssef NH, Elshahed MS (2020) Assessing anaerobic gut fungal diversity in herbivores using D1/D2 large ribosomal subunit sequencing and multi-year isolation. Environ Microbiol.https://doi.org/10.1111/1462-2920.15164
Gruninger RJ, Nguyen TTM, Reid ID, Yanke JL, Wang P, Abbott DW, Tsang A, McAllister T (2018) Application of transcriptomics to compare the carbohydrate active enzymes that are expressed by diverse genera of anaerobic fungi to degrade plant dell wall carbohydrates. Front Microbiol 9:1581. https://doi.org/10.3389/fmicb.2018.01581
Acknowledgements
The authors acknowledge Prof. Dr. Anna Schnürer for the support of barcode preparation and the 16s and ITS1 sequencing at the Department of Molecular Sciences, Swedish University of Agricultural Sciences, Sweden; the Freiburg Galaxy Team: Person X and Prof. Dr. Rolf Backofen for the website of metagenomic data analysis, Bioinformatics, University of Freiburg, Germany, funded by Collaborative Research Centre 992 Medical Epigenetics (DFG grant SFB 992/1 2012) and German Federal Ministry of Education and Research (BMBF grant 031 A538A de. NBI-RBC); the laboratory facility from Excellent Center of Waste Utilization and Management (ECoWaste) at King Mongkut’s University of Technology Thonburi, Thailand.
Funding
The authors gratefully acknowledge the financial support by the Petchra Pra Jom Klao Doctoral Scholarship from King Mongkut’s University of Technology Thonburi (KMUTT) awarded to Ms. Nitiya Thongbunrod. Research funding was supported by the Fundamental Fund (FF) of Thailand Science Research and Innovation (TSRI) and Ministry of Higher Education, Science, Research, and Innovation (MHESI).
Author information
Authors and Affiliations
Contributions
Pawinee Chaiprasert: conceptualization, project administration, funding acquisition, resources, supervision, and writing—reviewing and editing. Nitiya Thongbunrod: methodology, data curation, formal analysis, visualization, investigation, and writing—original draft preparation.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Thongbunrod, N., Chaiprasert, P. Potential of enriched and stabilized anaerobic lignocellulolytic fungi coexisting with bacteria and methanogens for enhanced methane production from rice straw. Biomass Conv. Bioref. 14, 8229–8250 (2024). https://doi.org/10.1007/s13399-022-03129-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13399-022-03129-1