Abstract
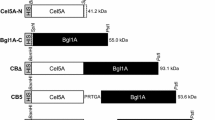
Primary cell walls and middle lamella of higher plants include homogalacturonan, the substrate for polygalacturonases. Seeking to enhance lignocellulose saccharification, chimeras between the endopolygalacturonase I from Chondrostereum (Stereum) purpureum (EndoPG-I) and family 3, 44, and 77 carbohydrate-binding modules (CBMs) from Hungateiclostridium thermocellum (Ht) or Ruminococcus flavefaciens (Rf) were constructed, expressed, and characterized. Chimeras presented similar KM values and pH/temperature optima as unfused EndoPG-I against citrus pectin, kcat/KM values 1.6, 1.7, and 1.3-fold higher for the HtCBM3-EndoPG, HtCBM44-EndoPG, and RtCBM77-EndoPG, respectively. Commercial enzyme cocktail supplementation with HtCBM44-EndoPG and RtCBM77-EndoPG increased reducing sugar release from untreated sugarcane bagasse by 35 and 25%, respectively. All chimeras increased reducing sugar release by 20–25% against orange bagasse compared with EndoPG-I or equimolar EndoPG-I/CBM mixtures. These results show that proximity between plant cell wall components in situ influences CBM-enzyme chimera activity and improves saccharification of lignocellulosic materials.



Similar content being viewed by others
Data availability
Not applicable.
Code availability
Not applicable.
References
IPCC, Masson-Delmotte V, Zhai P et al (2021) Climate Change 2021: The physical science basis. Contribution of Working Group I to the Sixth Assessment Report of the Intergovernmental Panel on Climate Change
Adiletta G, Brachi P, Riianova E et al (2020) A simplified biorefinery concept for the valorization of sugar beet pulp: ecofriendly isolation of pectin as a step preceding torrefaction. Waste Biomass Valoriz 11:2721–2733. https://doi.org/10.1007/S12649-019-00582-4/FIGURES/4
Kumar R, Tabatabaei M, Karimi K, Horváth IS (2016) Recent updates on lignocellulosic biomass derived ethanol - a review. Biofuel Res J 3:347–356. https://doi.org/10.18331/BRJ2016.3.1.4
Patel A, Shah AR (2021) Integrated lignocellulosic biorefinery: Gateway for production of second generation ethanol and value added products. J Bioresour Bioprod 6:108–128. https://doi.org/10.1016/J.JOBAB.2021.02.001
Loix C, Huybrechts M, Vangronsveld J et al (2017) Reciprocal interactions between cadmium-induced cell wall responses and oxidative stress in plants. Front Plant Sci 8:1867. https://doi.org/10.3389/FPLS.2017.01867/BIBTEX
Dranca F, Oroian M (2018) Extraction, purification and characterization of pectin from alternative sources with potential technological applications. Food Res Int 113:327–350. https://doi.org/10.1016/J.FOODRES.2018.06.065
Edwards MC, Doran-Peterson J (2012) Pectin-rich biomass as feedstock for fuel ethanol production. Appl Microbiol Biotechnol 95:565–575. https://doi.org/10.1007/S00253-012-4173-2/TABLES/5
Latarullo MBG, Tavares EQP, Maldonado GP et al (2016) Pectins, endopolygalacturonases, and bioenergy. Front Plant Sci 7:1401. https://doi.org/10.3389/FPLS.2016.01401/BIBTEX
Kameshwar AKS, Qin W (2018) Structural and functional properties of pectin and lignin–carbohydrate complexes de-esterases: a review. Bioresour Bioprocess 5:1–16. https://doi.org/10.1186/S40643-018-0230-8/FIGURES/5
Lombard V, GolacondaRamulu H, Drula E et al (2014) The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic acids research 42. https://doi.org/10.1093/NAR/GKT1178
Shimizu T, Miyairi K, Okuno T (2000) Determination of glycosylation sites, disulfide bridges, and the C-terminus of Stereum purpureum mature endopolygalacturonase I by electrospray ionization mass spectrometry. Eur J Biochem 267:2380–2389. https://doi.org/10.1046/J.1432-1327.2000.01249.X
Shimizu T, Shibata H, Araya T et al (2005) Expression, purification, and crystallization of endopolygalacturonase from a pathogenic fungus, Stereum purpureum, in Escherichia coli. Protein Expr Purif 44:130–135. https://doi.org/10.1016/J.PEP.2005.06.001
Hasui Y, Fukui Y, Kikuchi J et al (1998) Isolation, characterization, and sugar chain structure of endoPG Ia, Ib and Ic from Stereum purpureum. Biosci Biotechnol Biochem 62:852–857. https://doi.org/10.1271/BBB.62.852
Ogawa S, Shimizu T, Ohki H et al (2009) Expression, purification, and analyses of glycosylation and disulfide bonds of Stereum purpureum endopolygalacturonase I in Pichia pastoris. Protein Expr Purif 65:15–22. https://doi.org/10.1016/J.PEP.2008.12.014
Shimizu T, Nakatsu T, Miyairi K et al (2002) Active-site architecture of endopolygalacturonase I from Stereum purpureum revealed by crystal structures in native and ligand-bound forms at atomic resolution. Biochemistry 41:6651–6659. https://doi.org/10.1021/BI025541A
Ogawa S, Shimizu T, Kimura T et al (2010) The pro-form of Stereum purpureum endopolygalacturonase I is inactivated by a pro-sequence in the C-terminal region. Biosci Biotechnol Biochem 74:558–562. https://doi.org/10.1271/BBB.90754
Furtado GP, Carli S, Meleiro LP et al (2021) Enhanced hydrolytic efficiency of an engineered CBM11-glucanase enzyme chimera against barley β-d-glucan extracts. Food Chem 365:130460. https://doi.org/10.1016/J.FOODCHEM.2021.130460
Reddy Chichili VP, Kumar V, Sivaraman J (2013) Linkers in the structural biology of protein–protein interactions. Protein Sci 22:153. https://doi.org/10.1002/PRO.2206
Singh RK, Tiwari MK, Singh R, Lee JK (2013) From protein engineering to immobilization: promising strategies for the upgrade of industrial enzymes. Int J Mol Sci 14:1232. https://doi.org/10.3390/IJMS14011232
Hervé C, Rogowski A, Blake AW et al (2010) Carbohydrate-binding modules promote the enzymatic deconstruction of intact plant cell walls by targeting and proximity effects. Proc Natl Acad Sci U S A 107:15293–15298. https://doi.org/10.1073/PNAS.1005732107/-/DCSUPPLEMENTAL
Zhang X, Rogowski A, Zhao L et al (2014) Understanding how the complex molecular architecture of mannan-degrading hydrolases contributes to plant cell wall degradation *. J Biol Chem 289:2002–2012. https://doi.org/10.1074/JBC.M113.527770
Santos CR, Paiva JH, Sforça ML et al (2012) Dissecting structure-function-stability relationships of a thermostable GH5-CBM3 cellulase from Bacillus subtilis 168. Biochem J 441:95–104. https://doi.org/10.1042/BJ20110869
Furtado GP, Santos CR, Cordeiro RL et al (2015) Enhanced xyloglucan-specific endo-β-1,4-glucanase efficiency in an engineered CBM44-XegA chimera. Appl Microbiol Biotechnol 99:5095–5107. https://doi.org/10.1007/S00253-014-6324-0
Quan J, Tian J (2014) Circular polymerase extension cloning. Methods Mol Biol 1116:103–117. https://doi.org/10.1007/978-1-62703-764-8_8
Glynou K, Ioannou PC, Christopoulos TK (2003) One-step purification and refolding of recombinant photoprotein aequorin by immobilized metal-ion affinity chromatography. Protein Expr Purif 27:384–390. https://doi.org/10.1016/S1046-5928(02)00614-9
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. https://doi.org/10.1038/227680a0
Read SM, Northcote DH (1981) Minimization of variation in the response to different proteins of the Coomassie blue G dye-binding assay for protein. Anal Biochem 116:53–64. https://doi.org/10.1016/0003-2697(81)90321-3
Tomme P, Boraston A, Kormos JM et al (2000) Affinity electrophoresis for the identification and characterization of soluble sugar binding by carbohydrate-binding modules. Enzyme Microb Technol 27:453–458. https://doi.org/10.1016/S0141-0229(00)00246-5
Micsonai A, Wien F, Bulyáki É et al (2018) BeStSel: a web server for accurate protein secondary structure prediction and fold recognition from the circular dichroism spectra. Nucleic Acids Res 46:W315–W322. https://doi.org/10.1093/NAR/GKY497
McIlvaine TC (1921) A buffer solution for colorimetric comparison. J Biol Chem 49:183–186. https://doi.org/10.1016/S0021-9258(18)86000-8
Miller GL (1959) Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem 31:426–428. https://doi.org/10.1021/AC60147A030
Leone FA, Baranauskas JA, Furriel RPM, Borin IA (2005) SigrafW: an easy-to-use program for fitting enzyme kinetic data. Biochem Mol Biol Educ 33:399–403. https://doi.org/10.1002/BMB.2005.49403306399
de Souza AP, Leite DCC, Pattathil S et al (2013) Composition and structure of sugarcane cell wall polysaccharides: implications for second-generation bioethanol production. Bioenergy Res 6:564–579. https://doi.org/10.1007/S12155-012-9268-1/FIGURES/6
Carli S, Salgado JCS, Meleiro LP, Ward RJ (2021) Covalent immobilization of Chondrostereum purpureum endopolygalacturonase on ferromagnetic nanoparticles: catalytic properties and biotechnological application. Appl Biochem Biotechnol. https://doi.org/10.1007/S12010-021-03688-5
Carli S, Meleiro LP, Ward RJ (2019) Biochemical and kinetic characterization of the recombinant GH28 Stereum purpureum endopolygalacturonase and its biotechnological application. Int J Biol Macromol 137:469–474. https://doi.org/10.1016/J.IJBIOMAC.2019.06.165
Laskowski RA, Jabłońska J, Pravda L et al (2018) PDBsum: structural summaries of PDB entries. Protein Sci 27:129–134. https://doi.org/10.1002/PRO.3289
Grishina IB, Woody RW (1994) Contributions of tryptophan side chains to the circular dichroism of globular proteins: exciton couplets and coupled oscillators. Faraday Discuss 99:245–262. https://doi.org/10.1039/FD9949900245
Cattaneo C, Cesaro P, Spertino S et al (2018) Enhanced features of Dictyoglomus turgidum cellulase A engineered with carbohydrate binding module 11 from Clostridium thermocellum. Sci Rep 8:4402. https://doi.org/10.1038/s41598-018-22769-w
Fonseca-Maldonado R, Meleiro LP, Mendes LFS et al (2017) Lignocellulose binding of a Cel5A-RtCBM11 chimera with enhanced β-glucanase activity monitored by electron paramagnetic resonance. Biotechnol Biofuels 10:1–11. https://doi.org/10.1186/S13068-017-0964-0/FIGURES/5
Akhani RC, Patel AT, Patel MJ et al (2017) Ricin super family carbohydrate binding module 13 containing pectate lyase 1B from Bacillus licheniformis display hyper thermal stability. Asian J Biochem 12:36–43. https://doi.org/10.3923/AJB.2017.36.43
Selvendran RR, Ryden P (1990) Isolation and analysis of plant cell walls. Methods Plant Biochem 2:549–579. https://doi.org/10.1016/B978-0-12-461012-5.50022-7
Ajuong EMA, Breese MC (1998) Fourier transform infrared characterization of Pai wood (Afzelia africana Smith) extractives. Holz als Roh- und Werkstoff 56:139–142. https://doi.org/10.1007/S001070050285
Neves PV, Pitarelo AP, Ramos LP (2016) Production of cellulosic ethanol from sugarcane bagasse by steam explosion: effect of extractives content, acid catalysis and different fermentation technologies. Bioresour Technol 208:184–194. https://doi.org/10.1016/J.BIORTECH.2016.02.085
Arantes V, Saddler JN (2010) Access to cellulose limits the efficiency of enzymatic hydrolysis: the role of amorphogenesis. Biotechnol Biofuels 3:1–11. https://doi.org/10.1186/1754-6834-3-4/FIGURES/2
Li J, Zhou P, Liu H et al (2014) Synergism of cellulase, xylanase, and pectinase on hydrolyzing sugarcane bagasse resulting from different pretreatment technologies. Biores Technol 155:258–265. https://doi.org/10.1016/J.BIORTECH.2013.12.113
Delabona PdaS, Cota J, Hoffmam ZB et al (2013) Understanding the cellulolytic system of Trichoderma harzianum P49P11 and enhancing saccharification of pretreated sugarcane bagasse by supplementation with pectinase and α-L-arabinofuranosidase. Bioresour Technol 131:500–507. https://doi.org/10.1016/J.BIORTECH.2012.12.105
Thite VS, Nerurkar AS (2018) Physicochemical characterization of pectinase activity from Bacillus spp. and their accessory role in synergism with crude xylanase and commercial cellulase in enzyme cocktail mediated saccharification of agrowaste biomass. J Appl Microbiol 124:1147–1163. https://doi.org/10.1111/JAM.13718
Awan AT, Tsukamoto J, Tasic L (2013) Orange waste as a biomass for 2G-ethanol production using low cost enzymes and co-culture fermentation. RSC Adv 3:25071–25078. https://doi.org/10.1039/C3RA43722A
Cypriano DZ, da Silva LL, Tasic L (2018) High value-added products from the orange juice industry waste. Waste Manage 79:71–78. https://doi.org/10.1016/J.WASMAN.2018.07.028
Lionetti V, Francocci F, Ferrari S et al (2010) Engineering the cell wall by reducing de-methyl-esterified homogalacturonan improves saccharification of plant tissues for bioconversion. Proc Natl Acad Sci 107:616–621. https://doi.org/10.1073/PNAS.0907549107
Gama R, van Dyk JS, Pletschke BI (2015) Optimisation of enzymatic hydrolysis of apple pomace for production of biofuel and biorefinery chemicals using commercial enzymes. 3 Biotech 5:1075. https://doi.org/10.1007/S13205-015-0312-7
Acknowledgements
We thank André Justino for technical assistance.
Funding
This work was supported by the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) grants 2017/13734–3 (SC), 2016/17582–0 (LPM), 2019/21989–7 (JCSS), 2016/24139–6 (RJW), CNPq grant 305788/2017–5 (RJW) and the National Institute of Science and Technology of Bioethanol (INCT-Bioethanol) (FAPESP 2011/57908–6 and 2014/50884–5, CNPq 574002/2008–1 and 465319/2014–9).
Author information
Authors and Affiliations
Contributions
Sibeli Carli, data curation, formal analysis, investigation, methodology, figure preparation, visualization, and writing—original draft; Luana Parras Meleiro, data curation, formal analysis, investigation, methodology, figure preparation, visualization, and writing—review and editing; José Carlos Santos Salgado, data curation, formal analysis, investigation, methodology, figure preparation, visualization, and writing—original draft; and Richard John Ward, conceptualization, funding acquisition, project administration, resource management, team supervision, and writing—review and editing.
Corresponding author
Ethics declarations
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Carli, S., Parras Meleiro, L., Salgado, J.C.S. et al. Synthetic carbohydrate-binding module-endogalacturonase chimeras increase catalytic efficiency and saccharification of lignocellulose residues. Biomass Conv. Bioref. 14, 6369–6380 (2024). https://doi.org/10.1007/s13399-022-02716-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13399-022-02716-6