Abstract
This study investigated a chromosome hybrid zone between two chromosomal races of the common shrew (Sorex araneus). Gene flow and genetic structure of the hybrid zone, located in the northeast of Poland, were studied using seven polymorphic autosomal microsatellite loci (L9, L14, L33, L45, L67, L68, L97) and a Y-linked microsatellite locus (L8Y). Seventy-five animals (46 of the Łęgucki Młyn race and 29 of the Popielno race) from nine different localities were examined and the data were analyzed using hierarchical AMOVA and F-statistic. The studied microsatellite loci and races (divided into nine geographical populations) were characterized by observed heterozygosity (H O), expected heterozygosities within (H S), and between (H T) populations, inbreeding coefficient (F IS), fixation index (F ST), and average allelic richness (A). We found that genetic structuring within and between the two chromosome races were weak and non-significant. This finding and unconstrained gene flow between the races indicates a high level of migration within the Łęgucki Młyn/Popielno hybrid zone, suggesting that evolutionarily important genetic structuring does not occur in interracial zones where races which are not genetically distinct come into contact.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The common shrew (Sorex araneus), a small insectivore, seems to be an ideal object for the study of evolutionary processes, especially to investigate the role of chromosomes in speciation. It is a unique species, first of all, due to its great chromosome variation (both within and between populations) (Searle and Wójcik 1998). The basis of the quantitative and qualitative variation of the common shrew’s autosomes is chromosome aberrations in which the main cause is believed to be centric fusions (Robertsonian fusions) (Searle and Wójcik 1998). However, the recent study by White et al. (2010) taking into account different types of mutation (fusions, fissions, whole-arm reciprocal translocations, and zonal raciation events) revealed that fusions may be much less important than previously imagined.
As an effect of karyotype variation, S. araneus has been divided into almost 70 chromosome races, ten of which are found in Poland (Wójcik et al. 2003; Fedyk et al. 2008). In many cases, the neighboring races created hybrid zones, and several of them have already been described (Searle and Wójcik 1998; Jadwiszczak 2002; Moska 2003). In hybrid zones, detailed analysis has been done of either modifications enabling gene flow between neighboring races (acrocentric peak, recombinant peak, or elimination of acrocentrics from the zone) or factors responsible for the formation of isolation barriers between races (Searle and Wójcik 1998; Fredga and Narain 2000; Moska 2003).
Different molecular markers have been used in studies of the evolutionary processes of S. araneus (allozymes, mtDNA, microsatellites), of which microsatellites have been found a sensitive measure to detect patterns of hybridization. Up to now, the presence of over 40 highly polymorphic autosomal microsatellite loci and one polymorphic Y-chromosome microsatellite (L8Y) of this species have been reported (Wyttenbach et al. 1999; Balloux et al. 2000; Lugon-Moulin et al. 2000; Brünner et al. 2002a, b; Lugon-Moulin and Hausser 2002; Andersson 2004; Basset et al. 2006). L8Y provides an opportunity for investigating the level of male gene flow between populations and comparing it with the gene flow of both sexes (Lugon-Moulin and Hausser 2002).
The aim of the present study was to analyze the level of genetic differentiation in the Łęgucki Młyn/Popielno hybrid zone using microsatellite markers. Moreover, by analyzing the genetic structure within and between chromosome races, we tried to contribute to efforts of elucidating the role of interracial hybrid zones in the evolutionary process of speciation.
Material and methods
Study area
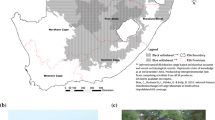
The hybrid zone between the Łęgucki Młyn and Popielno karyotypic races is located in the northeast of Poland (Fig. 1). The Łęgucki Młyn race (Łg) is characterized by chromosomes: g/r, h/k, i/o, j/l, m/n, p, q while the Popielno race (Po) by: g/r, h/q, i/k, j/l, m/n, o, p (Wójcik 1986; Moska 2003).
The location of the sampling sites in the Łęgucki Młyn/Popielno hybrid zone: P Prawdowo (53°48′N, 21°32′E), LK Lisunie-Kulinowo (53°46′N, 21°33′E), L Lipowo (53°47′N, 21°25′E), NM Nowy Most (53°44′N, 21°31′E), KI Krutyń I (53°42′N, 21°26′E), KII Krutyń II (53°41′N, 21°26′E), ZL Zielony Lasek (53°40′N, 21°27′E), Z Zakręt (53°39′N, 21°26′E), KM Karwica Mazurska (53°37′N, 21°27′E)
Shrews were collected over the period 1996–1999 from nine sites: Prawdowo (P), Lisunie-Kulinowo (LK), Lipowo (L), Nowy Most (NM), Krutyń I (KI), Krutyń II (KII), Zielony Lasek (ZL), Zakręt (Z), and Karwica Mazurska (KM) (Fig.1). Sites P, LK, L, NM, and ZL were situated in the area of the Łęgucki Młyn race (southern side of the hybrid zone) and KI, KII, Z, and KM in the area of the Popielno race (northern side of the hybrid zone) (Moska 2003).
Sampling and molecular methods
For the microsatellite analysis, 75 individuals of two races were used, 46 of the Łęgucki Młyn race and 29 of the Popielno race. Seven polymorphic autosomal microsatellites (L9, L14, L33, L45, L67, L68, L97) and a Y-linked microsatellite (L8Y) were used for the analysis. Amplification conditions for each locus were carried out according to Balloux et al. (1998) and Lugon-Moulin et al. (2000). In order to improve estimation of the size of the amplification product, one primer of each pair was labeled with a fluorescent dye (FAM or JOE) on the 5′-end, which allowed analyses with the use of an ABI 3100 Avant automated sequencer (Applied Biosystems).
Statistical analysis
The data were statistically analyzed with the application of two different approaches to the genetic structure of the Łęgucki Młyn/Popielno hybrid zone:
-
1.
First, the samples from the nine sites were considered as nine geographical populations;
-
2.
Second, the nine geographical populations were grouped according to their location in the area occupied. Populations P, LK, L, NM, and ZL situated in the area of the Łęgucki Młyn race formed group I; populations KI, KII, Z, and KM from the area of the Popielno race formed group II.
For the geographical populations, allelic richness (A), the observed (H O), and expected (H E) heterozygosity were computed for all loci using SAS v. 8.0 (User’s Guide 2000) and ARLEQUIN v. 3.1 (Excoffier et al. 2005).
Inbreeding coefficients (F IS) were estimated to indicate the within-population heterozygote deficiency due to non-random mating. F IS was calculated per locus, overall loci and per population for the geographical populations. Fixation index (F ST) was estimated per locus and for overall microsatellite loci. Permutations were used for testing the values of F IS and F ST for significant departure from zero. These tests were done using 10.000 permutations of alleles within samples (F IS) and 10.000 permutations of genotypes among samples for F ST (Arlequin v. 3.1).
Micro-Checker program v. 2.2.3 (Van Oosterhout et al. 2004) was used to identify genotyping errors due to null alleles.
To estimate the gene flow between the races, conventional F-statistics were used (Weir and Cockerham 1984). These statistics are recommended rather than R-statistics for studies with small sample sizes (ten individuals or less) and limited numbers of loci (less than 20) (Gaggiotti et al. 1999).
The genetic structure of populations was calculated using the analysis of molecular variance (AMOVA) as implemented in Arlequin v. 3.1. A hierarchical analysis of variance divided the total variance into different parts of covariance which were then used to calculate different fixation indices (Excoffier 2000). Significance levels of the different fixation indices were obtained by permuting genotypes for F ST, among geographical populations; for F SC, among geographical populations within the races; and for F CT, among chromosomal races (Excoffier et al. 1992).
Results
Polymorphism and genetic variability
Most autosomal loci displayed a high level of polymorphism. One hundred and thirty six alleles were found at seven autosomal microsatellite loci. The most variable locus (L97) had 30 alleles, whereas the least variable one (L67) had 12 alleles (Table 1).
The average allelic richness (A) in the different geographical populations ranged from 2.75 to 10.11, and was smallest in population KM and greatest in population Z (Table 2). Observed heterozygosities (H O) per locus ranged from 0.57 to 0.86, and over all loci amounted to 0.75. Expected heterozygosities within samples (H S) ranged from 0.68 to 0.83 with an average value of 0.76, whereas over all samples (H T) ranged from 0.73 to 0.94 with the average value of 0.87 (Table 1). The average H O in different geographical populations ranged from 0.64 to 0.86, whereas H E fluctuated from 0.75 to 0.89 (Table 2).
Genetic structure of the Łęgucki Młyn/Popielno hybrid zone
FIS estimated over all loci revealed significant departure from HWE expectations (FIS = 0.063, P < 0.01). The FIS observed in each geographical population ranged from 0.12 (KI) to 0.29 (KM) and departed significantly from HWE in two of them: L/K and L (P < 0.01; Table 2). All other geographical populations showed no FIS values significantly different from zero. In the analysis of mean FIS over all loci, a very high and significant heterozygote deficit (FIS = 0.13) was observed. FIS per locus over all populations ranged from 0.00 (L68) to 0.28 (L45). Results from the permutation procedure indicated that the FIS values were significant for three loci: L33, L45, and L97 (Table 1).
The F ST over all geographical populations was small and not significant (F ST = 0.02; Table 1). Locus-specific F ST ranged from 0.008 (L68) to 0.038 (L97), and significant genetic differentiation was found only at locus L97 (Table 1). The mean F ST was also calculated for both races separately. The Łęgucki Młyn race (populations P, L/K, L, NM, ZL) showed F ST of 0.027, whereas the Popielno race showed F ST of 0.003. For both races values of F ST were small and non-significant (Table 1).
The AMOVA analyses based on F ST showed a lack of genetic differentiation between chromosomal races, both for autosomal microsatellite loci and for the Y-locus. For the autosomal loci, 0% (−0.26%) of the total variation was caused by variation between races, whereas for L8Y it was 3.03% of the total variation. Most of the variation was found within geographical populations as regards to both autosomal loci and L8Y (99% and 90%, respectively). For all genetic markers low and non-significant variation was found among geographical populations within the races.
Discussion
So far, detailed studies using DNA microsatellites have been carried out in three hybrid zones: two located in Sweden (Uppsala/Hällefors and Abisko/Sidensjö) (Wyttenbach et al. 1999; Andersson 2004), and one located in Poland (Drnholec/Białowieża) (Jadwiszczak et al. 2006). The Abisko/Sidensjö and Drnholec/Białowieża hybrid zones are the ones where hybridization of karyotypically divergent chromosome races takes place (races belong to distinct karyotypic groups: NEKG and WEKG, WEKG and EEKG, respectively) (Fredga 1996; Jadwiszczak 2002). The analysis of genetic differentiation revealed weak genetic structuring and in consequence unimpeded gene flow between the races. However, in the Abisko/Sidensjö hybrid zone, significant reduction of male gene flow was found when the genetic marker linked to Y-chromosome (L8Y) was studied (Andersson 2004; Jadwiszczak et al. 2006). In the Uppsala/Hällefors hybrid zone, both races are not chromosomally distinct (they belong to the same evolutionary group, WEKG) (Narain and Fredga 1996). Based on genetic differentiation of autosomal microsatellite loci, weak genetic structuring over all localities was found (Wyttenbach et al. 1999).
A comparison of the studied hybrid zones leads to the conclusion that the Łęgucki Młyn/Popielno hybrid zone strongly resembles the Uppsala/Hällefors hybrid zone. These two races (Łg and Po) are chromosomally quite closely related (both races represent EEKG) and in the hybrid zone complex heterozygotes are theoretically expected to form only short chains (CIV or CV) during meiosis.
Molecular analysis of seven autosomal loci did not reveal significant differentiation between the Łęgucki Młyn and Popielno chromosomal races. The F ST values were small and not significant, either over all geographical populations or within the races. Our results were comparable with the weak genetic structuring observed in the similar Uppsala/Hällefors hybrid zone.
When we analyzed the level of genetic structure with regard to the Y-chromosome microsatellite, we found a level ten times higher compared to autosomal loci (F ST = 0.10). Our findings are comparable to results previously reported by different authors who observed differences in the estimates of genetic structure based on the Y-microsatellite variation compared to the autosomal microsatellite (Andersson et al. 2004; Balloux et al. 2000).
In our study, F IS values over all loci and over all populations were high and significantly different from zero (F IS = 0.13 and F IS = 0.06, respectively). A closer look at F IS for each geographical population revealed that populations LK and L showed a high and significant heterozygote deficit (F IS = 0.14 and F IS = 0.10, respectively; P < 0.01). The high F IS may be partly cased by the Wahlund effect (White and Searle 2007). In one geographical population (LK), shrews were caught on two meadows separated by a busy road, so a deficit of heterozygotes could be caused by pooling independent populations in HWE. Previous studies have shown no indications of null alleles at any of the loci used in this study, indicating that high F IS was probably not caused by presence of null alleles (Wyttenbach and Hausser 1996; Wyttenbach et al. 1999; White and Searle 2007). However, the analysis of the data used in our study with the use of Micro-Checker software identified null alleles in three loci (L33, L45, and L97) where a significant (P < 0.01) inbreeding coefficient was estimated. This is probably the most reasonable explanation of the lack of heterozygosity in those loci. The deficit of heterozygotes observed in our study may be also explained by the small sample sizes (particularly from populations L, KII, and KM, where samples of fewer than five individuals were taken).
In conclusion, the results described in this paper give insight into the speciation process which may take place in hybrid zones. Our study, with the use of microsatellite markers, did not reveal significant differences between the genetically close Łęgucki Młyn and Popielno chromosomal races, which was proven by small chromosomal differentiation. It seems that the evolutionarily important genetic structuring does not occur in interracial zones, where races which are not genetically distinct come into contact.
References
Andersson A-C (2004) Postglacial population history of the common shrew (Sorex araneus) in Fennoscandia. Ph.D. Thesis, Uppsala University
Andersson A-C, Narain Y, Tegelström H, Fredga K (2004) No apparent reduction of gene flow in a hybrid zone between the West and North European karyotypic groups of the common shrew, Sorex araneus. Mol Ecol 13:1205–1215. doi:10.1111/j.1365-294X.2004.02146.x
Balloux F, Ecoffey E, Fumagalli L, Goudet J, Wyttenbach A, Hausser J (1998) Microsatellite conservation, polymorphism, and GC content in shrews of the genus Sorex (Insectivora, Mammalia). Mol Biol Evol 15:473–475
Balloux F, Lugon-Moulin N, Hausser J (2000) Estimating gene flow across hybrid zones: how reliable are microsatellites? Acta theriol 45(suppl 1):93–101
Basset P, Yannic G, Yang F, O'Brien PCM, Graphodatsky AS, Ferguson-Smith MA, Balmus G, Volobouev VT, Hausser J (2006) Chromosome localization of microsatellite markers in the shrews of the Sorex araneus group. Chromosome Res 14:253–262. doi:10.1007/s10577-006-1041-x
Brünner H, Lugon-Moulin N, Hausser J (2002a) Alps, genes, and chromosomes: their role in the formation of species in the Sorex araneus group (Mammalia, Insectivora), as inferred from two hybrid zones. Cytogenet Genome Res 96:85–96
Brünner H, Lugon-Moulin N, Balloux F, Fumagalli L, Hausser J (2002b) A taxonomical re-evaluation of the Valais chromosome race of the common shrew Sorex araneus (Insectivora: Soricidae). Acta Theriol 47:245–275
Excoffier L (2000) Analysis of population subdivision. In: Balding D, Bishop M, Cannings C (eds) Handbook of statistical genetics. Wiley and Sons, New York, NY. doi:10.1002/0470022620.bbc25
Excoffier L, Smouse P, Quattro J (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial restriction date. Genetics 131:479–491
Excoffier L, Laval G, Schneider S (2005) Arlequin ver. 3.0: an integrated software package for population genetics data analysis. Evol Bioinfo Online 1:47–50
Fedyk S, Wójcik JM, Chętnicki W, Mączewski S (2008) Invalidation of Stobnica chromosome race of the common shrew, Sorex araneus. Acta Theriol 53:375–380
Fredga K (1996) The chromosome races of Sorex araneus in Scandinavia. Hereditas 125:123–135. doi:10.1111/j.1601-5223.1996.00123.x
Fredga K, Narain Y (2000) The complex hybrid zone between Abisko and Sidensjö chromosome races of Sorex araneus in Sweden. Biol J Linn Soc 70:285–307. doi:10.1006/bijl.1999.0378
Gaggiotti OE, Lange O, Rassmann K, Gliddon C (1999) A comparison of two indirect methods for estimating average levels of gene flow using microsatellites data. Mol Ecol 8:1513–1520. doi:10.1046/j.1365-294x.1999.00730.x
Jadwiszczak KA (2002) Chromosome structure of the hybrid zone between the Drnholec and Białowieża races of the common shrew, Sorex araneus Linnaeus, 1758. Ph.D. Thesis, Białystok University. [In Polish]
Jadwiszczak KA, Ratkiewicz M, Banaszek A (2006) Analysis of molecular differentiation in a hybrid zone between chromosomally distinct races of the common shrew Sorex araneus (Insectivora: Soricidae) suggests their common ancestry. Biol J Linn Soc 89:79–90. doi:10.1111/j.1095-8312.2006.00659.x
Lugon-Moulin N, Balloux F, Hausser J (2000) Genetic differentiation of common shrew Sorex araneus populations among different alpine valleys revealed by microsatellites. Acta theriol 45(suppl 1):103–117
Lugon-Moulin N, Hausser J (2002) Phylogenetical structure, postglacial recolonization and barriers to gene flow in the distinctive Valais chromosome race of the common shrew (Sorex araneus). Mol Ecol 11:785–794
Moska M (2003) A hybrid zone between the chromosomal races Łęgucki Młyn and Popielno of the common shrew (Sorex araneus) in north-eastern Poland: preliminary results. Acta Theriol 48(4):441–455
Narain Y, Fredga K (1996) A hybrid zone between Hällefors and Uppsala chromosome races of Sorex araneus in central Sweden. Hereditas 125:137–145. doi:10.1111/j.1601-5223.1996.00137.x
User’s Guide SAS (2000) Version 8.0 Edition. SAS InstituteInc, Cary, NC, USA
Searle JB, Wójcik JM (1998) Chromosomal evolution: the case of Sorex araneus. In: Wójcik JM, Wolsan M (eds) Evolution of Shrews. Mammal Research Institute, Polish Academy of Sciences, Białowieża, pp 219–268
Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P (2004) MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite date. Mol Ecol Notes 4:535–538. doi:10.1111/j.1471-8286.2004.00684.x
Weir BC, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370
White TA, Searle JB (2007) Genetic diversity and population size: island populations of the common shrew, Sorex araneus. Mol Ecol 16:2005–2016. doi:10.1111/j.1365-294X.2007.03296.x
White TA, Bordewich M, Searle JB (2010) A network approach to study karyotypic evolution: the chromosomal races of the common shrew (Sorex araneus) and house mouse (Mus musculus) as model systems. Syst Biol 59(3):262–276. doi:10.1093/sysbio/syq004
Wójcik JM (1986) Karyotypic races of the common shrew (Sorex araneus L.) from northern Poland. Experientia 42:960–962. doi:10.1007/BF01941780
Wójcik JM, Borodin PM, Fedyk S, Fredga K, Hausser J, Mishta A, Orlov VN, Searle JB, Volobouev VT, Zima J (2003) The list of chromosome races of the common shrew Sorex araneus (updated 2002). Mammalia 68:169–178
Wyttenbach A, Hausser J (1996) The fixation of metacentric chromosomes 619 during the chromosomal evolution in the common shrew (Sorex araneus, Insectivora). Hereditas 125:209–217. doi:10.1111/j.1601-5223.1996.00209.x
Wyttenbach A, Narain Y, Fredga K (1999) Genetic structuring and gene flow in a hybrid zone between two chromosome races of the common shrew (Sorex araneus, Insectivora) revealed by microsatellites. Hereditas 82:79–88. doi:10.1038/sj.hdy.6884520
Acknowledgements
The field assistance of Professor S. Fedyk and Dr. W. Chętnicki is gratefully acknowledged. We wish to thank M. Kowalska and A. Jonkisz for their laboratory help. Thanks also go to anonymous referees for their useful comments.
Declaration concerning ethical standards
Our experiments comply with all current laws in Poland.
Conflict of interest
The authors declare that they have no conflict of interest.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by: Jan M. Wójcik
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Moska, M., Wierzbicki, H., Macierzyńska, A. et al. A microsatellite study in the Łęgucki Młyn/Popielno hybrid zone reveals no genetic differentiation between two chromosome races of the common shrew (Sorex araneus). Acta Theriol 56, 117–122 (2011). https://doi.org/10.1007/s13364-011-0029-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13364-011-0029-x