Abstract
We report a gas-phase UV photodissociation study investigating non-covalent interactions between neutral hydrophobic pentapeptides and peptide ions incorporating a diazirine-tagged photoleucine residue. Phenylalanine (Phe) and proline (Pro) were chosen as the conformation-affecting residues that were incorporated into a small library of neutral pentapeptides. Gas-phase ion-molecule complexes of these peptides with photo-labeled pentapeptides were subjected to photodissociation. Selective photocleavage of the diazirine ring at 355 nm formed short-lived carbene intermediates that underwent cross-linking by insertion into H–X bonds of the target peptide. The cross-link positions were established from collision-induced dissociation tandem mass spectra (CID-MS3) providing sequence information on the covalent adducts. Effects of the amino acid residue (Pro or Phe) and its position in the target peptide sequence were evaluated. For proline-containing peptides, interactions resulting in covalent cross-links in these complexes became more prominent as proline was moved towards the C-terminus of the target peptide sequence. The photocross-linking yields of phenylalanine-containing peptides depended on the position of both phenylalanine and photoleucine. Density functional theory calculations were used to assign structures of low-energy conformers of the (GLPMG + GLL*LK + H)+ complex. Born-Oppenheimer molecular dynamics trajectory calculations were used to capture the thermal motion in the complexes within 100 ps and determine close contacts between the incipient carbene and the H–X bonds in the target peptide. This provided atomic-level resolution of potential cross-links that aided spectra interpretation and was in agreement with experimental data.

ᅟ
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Chemical cross-linking relies on the formation of covalent bonds between components of a non-covalent complex or different sites in a single large molecule [1, 2]. With the introduction and development of photoactivated [2] and photodissociative [3,4,5] cross-linking reagents, it became possible to generate highly reactive and short-lived intermediates to map contacts between the transient reactive group and sites in the target molecule. The diazirine ring, in particular, has been utilized as a photocleavable group that undergoes specific elimination of nitrogen by irradiation at 355–370 nm [6]. Diazirine photolysis forms a transient reactive carbene that can undergo insertion into proximate X–H bonds, forming covalent cross-links (Scheme 1). Carbene insertion into C–H bonds has been shown to occur even at − 196 °C [7] and is thought to proceed without an energy barrier [8]. This makes carbenes excellent photoaffinity reagents that undergo indiscriminate insertion into C–H bonds of amino acid side chains upon contact [2]. In contrast, aliphatic carbenes are known to form short-lived ion-pair complexes with O–H bonds [9] that may introduce a positive bias towards insertions into polar groups in peptides and proteins [10].
The diazirine ring can be introduced into the protein or peptide of interest by using photolabile amino acid residues such as photoleucine (L-2-amino-4,4-azi-pentanoic acid, L*), photomethionine (L-2-amino-5,5-azi-hexanoic acid, M*) [11], or the recently reported photolysine (L-2,6-diamino-4,4-azi-hexanoic acid) [12]. Photoleucine has been shown to be a useful surrogate for the corresponding natural amino acid, and can be used for photochemical footprinting in expressed proteins [11].
Recently, photochemical cross-linking using diazirine tags has been applied to gas-phase peptide-peptide ion complexes [10]. In this approach, we used electrospray ionization to produce non-covalent ion-molecule complexes of a photo-tagged peptide with target peptides that were selected by mass and stored in an ion trap mass spectrometer. Irradiation with a 355-nm laser beam of the mass-selected complex resulted in N2 elimination that was associated with covalent bond formation in cross-links. Cross-linking in gas-phase peptide-peptide ion complexes has been shown to achieve efficiencies [10, 13] that exceeded those observed in solution cross-linking using diazirine-tagged peptides [14]. Recently, we have expanded the portfolio of diazirine-tagged amino acid residues for gas-phase peptide cross-linking by position-tunable tags placed at the ε-amino group in lysine residues and N-terminal amino groups [13]. Photodissociation at 157 nm of peptide complexes [15] and cation-anion reactions [16] have also been used to form covalent bonds in gas-phase ions.
Scheme 1 indicates that photolytically produced carbene intermediates can be quenched by competitive 1,2-hydrogen shifts from the neighboring methylene or methyl group, forming unreactive olefins. While these side reactions, as well as reactions with solvent, are detrimental in solution studies [17], the nanosecond kinetics of the 1,2-H shift [18] provides an internal clock for cross-linking in gas-phase complexes where insertions in the X–H bonds occur only if they proceed on a comparable time scale. We utilized this feature for determining the time scale for molecular dynamics calculations of trajectories describing the thermal motion in gas-phase complexes with the goal of identifying close contacts between the incipient carbene atom of the photo-tagged peptide and X–H bonds in the target peptide. Herein, we exploited our gas-phase cross-linking technique to study conformations of gas-phase ion-molecule complexes generated from the combination of photoactive peptides GL*LLK, GLL*LK, and GLLL*K with two small libraries of neutral hydrophobic pentapeptides. One library included proline-containing pentapeptides PGLMG, GPLMG, GLPMG, GLMPG, and GLMGP where the Pro residue was systematically moved along the peptide sequence. In the other library, Pro residues were replaced by Phe as in GFLMG, GLFMG, and GLMFG. Proline is known to affect the chain conformation in proteins [19,20,21,22,23] where it forms β turns, reversing the direction of the polypeptide chain [23]. Proline has a distinctive cyclic structure and tertiary amide nitrogen that distinguish it from other residues [21]. We hypothesized that the conformational properties of proline could potentially play a role in affecting the conformation of a small neutral peptide in the gas phase, and thus enhance the specificity of its interaction with photopentapeptides. Polyproline peptide ions have been shown by ion mobility to exhibit interesting conformational properties in the gas phase [24]. In addition to proline, we also considered phenylalanine as another residue for modulating non-covalent interactions of the neutral pentapeptide with its photopeptide counterpart. In contrast to proline, phenylalanine does not have a strong effect on the backbone conformation of the peptide sequence. However, the aromatic side chain could restrict the motion of the neutral Phe-containing peptides in the direction perpendicular to the ring plane, which is aligned with the peptide ion backbone. Pro- and Phe-containing peptides with the sequences used in this study appear as sequence motifs in a number of biologically important proteins. For example, the motif PGLMG has been found in over 1000 proteins present in the Boreoeutheria class [25]. The PGLMG motif appears mostly in type I through IV collagen proteins, which are the major structural components of the basement membrane. Hence, the insight gained from the gas-phase study could enhance our understanding of non-covalent binding in large biomolecules.
The size of the neutral target peptides was limited to five residues to allow us to conduct a computational analysis of conformational trajectories using Born-Oppenheimer molecular dynamics (BOMD) and compare the results to experimental data. All BOMD calculations were run at the PM6-D3H4 augmented semi-empirical level of quantum theory that accounts for hydrogen bonding and London dispersion interactions [26]. Both these interactions must be considered in addressing the stability and dynamics of the gas-phase complexes [10]. We wish to demonstrate the effect that proline and phenylalanine have on modulating non-covalent interactions of dimer complexes formed in the gas phase. Our other goal was to illustrate the utility of diazirine photochemistry in conjunction with BOMD computational analysis for probing the structure and dynamics of peptide-peptide ion interactions in the gas phase.
Experimental Section
Materials
Photo-labeled peptides GL*LLK, GLL*LK, and GLLL*K were synthesized using standard solid phase peptide Fmoc techniques on Wang resin (Bachem Americas, Torrance, CA, USA) [27, 28]. Photoleucine (L*, L-2-amino-4,4-azi-pentanoic acid) and its Fmoc-protected derivative were purchased from Life Technologies, Rockford, IL, USA. All neutral peptides, PGLMG, GPLMG, GLPMG, GMLPG, GLMGP, GFLMG, GLFMG, and GMLFG, were synthesized on a CEM Liberty Blue synthesizer (CEM, Matthews, NC, USA) using standard Fmoc techniques.
Methods
The peptide-peptide complexes were formed by electrospray of peptide mixture solutions. Typically, 50 μL of the neutral peptide solution (5–10 μM in 50:50:1 methanol/water/acetic acid) was mixed with 100 μL of the photopeptide solution (5–10 μM in 50:50:1 methanol/water/acetic acid). The mixed solution was electrosprayed at 2.2–2.3 kV from a pulled fused silica with a syringe pump capillary. The singly charged complex ions were mass selected, stored in a modified LTQ-XL ETD linear ion trap (LIT) mass spectrometer (Thermo Electron Fisher, San Jose, CA, USA), and then photodissociated at 355 nm [29]. Photodissociative loss of N2 formed product ions that were selected by mass and subjected to CID-MS3 for sequencing.
Briefly, the UV photodissociation-tandem mass spectrometry (UVPD-MS2) experiments were conducted by first isolating the singly charged complex ions of interest in the LIT for a period of time, and then irradiating them with the laser beam, which is produced by an EKSPLA NL 301 HT (Altos Photonics, Bozeman, MT, USA) Nd-YAG laser operating at 20 Hz frequency with a 3–6-ns pulse width. Due to the maximum frequency at which the laser is fired (20 Hz), each laser pulse requires a 50-ms ion storage time. The number of laser pulses could be varied to accomplish a pulse-dependent study by changing the storage time of the trapped ions in the LIT. A third harmonic frequency generator was also included in the path to produce a single 355-nm wavelength at a maximum 120 mJ/pulse peak power. However, a typical photodissociation experiment only used 18 mJ/pulse light intensity. Both the laser system and the LTQ-XL are set on an optical table for optimum alignment. The laser was interfaced to the LTQ by LabView software (National Instruments, Austin, TX, USA) that receives a signal from a TTL pulse on pin14 of the J1 connector on the LTQ console. Laser pulses are triggered internally by the EKSPLA system, but the power is controlled for each pulse by commands from the LabView software.
Born-Oppenheimer Molecular Dynamics Calculations
All molecular dynamics calculations used the all-valence-electron semiempirical PM6-D3H4 [26] method with the Berendsen thermostat algorithm [30] at a constant temperature of 310 K. The MD analysis was divided in two parts. The first part involved the mapping of the conformational space of the peptide complexes. Twenty-five initial structures were generated with different orientations of the photoactivable diazirine, and the charging proton was placed on the ε-amine group of the lysine residue in the photopentapeptide. For each initial structure, a preliminary MD was conducted with a duration of 20 ps to obtain 200 snapshots. The total of 25 × 200 = 5000 snapshot structures were then subjected to full optimization with PM6-D3H4 that was run by MOPAC coupled with the Cuby4 framework that provided a high-level platform [31, 32]. The 5000 fully optimized PM6-D3H4 snapshot structures were sorted out by energy, and 650 structures within a 50 kJ mol−1 window were re-optimized by density functional theory (DFT) calculations employing the BLYP functional [33, 34] and the def-SV(P) basis set [35] including dispersion corrections. These calculations were carried out using TurboMole software [36]. Twelve complexes with the lowest DFT energies, and having conformations with different orientations of the L* and target peptide subunits, were then selected and submitted as initial structures to a full BOMD run for 100 ps. The trajectories were run at 310 K, which was the ion trap temperature used in the measurements.
Thermochemical calculations were performed using the Gaussian 09 (Revision A.02.) [37] and Gaussian 16 (Revision A.03) [38] suites of programs. Twelve lowest-energy structures from the BLYP/def-SV(P) calculations were re-optimized with B3LYP/6-31G(d,p) to provide harmonic frequencies that were used to evaluate enthalpies, entropies, and thermal free-energy corrections at 310 K. In separate runs, these 12 initial structures were re-optimized with ωB97X-D [39, 40] and the 6–31 + G(d,p) basis set. These calculations included dispersion interactions in the complexes, and the obtained energies were used to evaluate the electronic terms of the free energies. The calculated relative free energies including electronic, vibrational, and rotational terms (Table S1, Supporting Information) were used to calculate equilibrium molar fractions of the gas-phase complexes at 310 K (Table S2, Supporting Information).
Note on the Complex Ion Nomenclature
To describe the fragment ions originating from the complexes, we utilize the nomenclature system previously described by Shaffer et al. [10]. Briefly, all the target peptides are referred to with a small letter (m) while the photopeptides are labeled with a capital letter (M), such as (mM – N2 + H)+ for complexes produced by loss of N2. Fragment ions resulting from backbone dissociation in the target peptide part of the complex are denoted as bnM (retaining target peptide N-terminal residues) and ynM (retaining target peptide C-terminal residues). Similarly, fragment ions resulting from backbone cleavage in the photopeptide part of the complex are denoted as mBm and mYm. Fragment ions combining backbone dissociations in both the neutral and photopeptide parts are labeled adequately as bnBm, ynYm, etc., according to the observed m/z. A simple representation for this ion nomenclature system is illustrated in Fig. 1 for the dimer complex (GLPMG + GLL*LK + H)+.
Results and Discussion
Photocross-Linking of Pro-Containing Target Peptides
Electrospray ionization (ESI) of peptide-peptide complexes yielded singly charged ions of the non-covalent complexes in 1.9–14.4% yields relative to the singly charged peptide ions that were the main species in the ESI mass spectra (Table 1). This is illustrated for the complex of GPLMG with GL*LLK at m/z 1028 (Fig. 2a).
The mass spectra of the other combinations of target and photoactive peptide complexes are shown in Figs. S1–S14 (Supporting Information). The (GPLMG + GL*LLK + H)+ complex was selected by mass and subjected to collision-induced dissociation (CID) and photodissociation at 355 nm (UVPD). CID-MS2 of (GPLMG + GL*LLK + H)+ resulted in complex dissociation, forming (GL*LLK + H)+ (m/z 555) as the major charged fragment along with its dissociation product, (GL*LLK – N2 + H)+ (m/z 527, Fig. 2b). CID-MS2 of the other complexes gave very similar results. These results indicated that the non-covalent interactions of the peptide moieties in the complexes were readily disrupted by CID without substantially affecting the photoactive diazirine group [10, 13]. In contrast, UVPD-MS2 resulted in a major loss of N2 while preserving the complex as an (mM – N2 + H)+ ion. This is illustrated by the UVPD-MS2 spectrum of (GPLMG + GL*LLK + H)+ displaying the (mM – N2 + H)+ ions at m/z 1000 (Fig. 2c). To achieve high (> 50%) conversion of the precursor ions, 36 laser pulses were uniformly used for UVPD-MS2. Note that the photodissociation products are transparent at 355 nm and do not undergo further depletion by photodissociation. In addition to loss of N2, UVPD-MS2 also resulted in the breakdown of the complex forming (M – N2 + H)+ ions, as shown for GL*LLK (Fig. 2c). The formation of (M – N2 + H)+ monomers upon UVPD was indicative of the fraction of complexes in which the photopeptide lost N2 forming an olefin without cross-linking to the target peptide. This mode of complex dissociation was consistent with the energetics of N2 elimination from L* followed by carbene isomerization to an olefin. The carbene to olefin conversion in L* has been calculated to be ca. 200 kJ mol−1 exothermic [41], thus providing internal energy to drive dissociation of the non-covalent complex.
The formation of the new covalent bond upon carbene insertion was detected by subjecting the [mM−N2 + H]+ ions to CID-MS3 for structural analysis by gas-phase sequencing. CID of [mM−N2 + H]+ ions revealed that the majority of fragment ions originated from covalent adducts, as indicated by the m/z values that were between that of (mM−N2 + H)+ and (M−N2 + H)+. This is illustrated by the CID-MS3 spectrum of (GPLMG + GL*LLK – N2 + H)+ (Fig. 3). The CID-MS3 spectra of the other proline peptide complexes are shown in Figs. S15–S28 (Supporting Information). The fragment ions were assigned on the basis of the known sequence of the photoactive and target peptides.
The photodissociative conversion of peptide-peptide ion complexes was quantified in two ways [10, 13]. First, the relative intensities of the total covalent and non-covalent ion fraction in the UVPD-MS2 spectra, (mM – N2 + H)+, were expressed as per cent ratios R(MS2) (Eq. 1):
where [mM – N2 + H]+ and [M – N2 + H]+ are the respective intensities of the denitrogenated dimer and its monomer dissociation product. The R(MS2) values for all studied combinations of proline target peptides and photopeptides exceeded 30% (Table 2). The sequence positions of the Pro and L* residues had only a moderate effect on R(MS2). As a general trend, moving the Pro residue from the N-terminus to the C-terminus resulted in a slightly increased R(MS2) for complexes with all three L*-tagged photopeptides (Table 2). The highest R(MS2) was observed for the (GMLPG + GLL*LK + H)+ complex (44%, Table 2).
Second, the fractions of cross-linked complexes were determined from CID-MS3 spectra of (mM – N2 + H)+ as a percent ratio of the sum of backbone fragment ion intensities (Fi) relative to all fragment ions including the (M – N2 + H)+ monomer (Eq. 2):
The R(MS3) values ranged between 64 and 85% and displayed different trends depending on the Pro and L* positions (Table 3), as discussed below. We note that the reported R(MS3) yields should be viewed as lower bounds of the actual cross-linked complexes for two reasons. First, including in R(MS3), the intensities of fragment ions formed by loss of ammonia and water from (mM – N2 + H)+ ions further increased the R(MS3) by as much as 20% to reach the high of 90% covalent cross-linking in the (GLPMG + GL*LLK + H)+ complex (Table 3). The fragment ion intensities due to the loss of ammonia depended on the target peptide sequence (Table 3). We did not attempt to interpret these effects because the origin of the ammonia molecule, i.e., from the photopeptide or target peptide, was unknown. Second, covalent bonds susceptible to dissociation, such as those in esters and amides formed by carbene insertion to carboxyl O–H and amide N–H bonds, respectively, may undergo CID cleavage, forming (M – N2 + H)+ fragment ions that would be counted as originating from non-covalent (mM – N2 + H)+ complexes. Carbene cross-linking to target peptide carboxyl group forming an ester linkage was discussed previously and found likely in some structures [10]. The combined yields, R(MS2) × R(MS3), were between 19 and 35% for the combinations of Pro target and L*-tagged peptides (Table 2).
Photocross-Linking of Phe-Containing Target Peptides
Electrospray ionization of solution mixtures composed of Phe-containing target peptides and photo-labeled peptides formed singly charged non-covalent dimer complexes at m/z 1078. The ESI yield of the formation of Phe-containing complexes was 2.2–12% relative to the singly charged monomers, as shown for (GFLMG + GL*LLK + H)+ (Fig. 4a). The non-covalent dimer complexes (m/z 1078) were selected by mass and subjected to CID-MS2 and UVPD-MS2 in a similar fashion as for the Pro-containing dimer complexes. CID-MS2 of Phe-containing dimers showed two main product ions, which were the (M – N2 + H)+ (m/z 527) and (m + H)+ (m/z 555) (Fig. 4b). UVPD-MS2 at 355 nm of the complexes resulted in the formation of (M – N2 + H)+ (m/z 527) and a major loss of N2, to form (mM – N2 + H)+ (m/z 1050) ions, as illustrated for the (GFLMG + GL*LLK + H)+ complex (Fig. 4c). The mass spectra of the other combinations of Phe-containing complexes showed similar results (Figs. S29–S36, Supporting Information). The dissociations of the complexes observed upon CID-MS2 indicated that the non-covalent interactions between the peptide components were broken under CID conditions and were not residue-specific. The formation of (mM – N2 + H)+ (m/z 1050) under UVPD conditions suggested the formation of a new covalent bond between the two monomers, and this was further probed by subjecting the (mM – N2 + H)+ ions to CID-MS3, as illustrated for (GFLMG + GL*LLK – N2 + H)+ (m/z 1050) (Fig. 5).
The R(MS2) values of all Phe-containing complexes were in the 31–38% range (Table 2), whereby the highest R(MS2) value was obtained when both the Phe and the L* residues were near the C-terminus. This indicated that there may exist an interaction between the π system of the aromatic ring of the Phe residue and the charge located on the ε-amine of the C-terminal lysine residue. This type of interaction could potentially help enhance the photocross-linking efficiency between the peptide units. The R(MS3) values of the Phe-containing complexes ranged within 65–80%, and were not much affected by including minor (mM – N2 – NH3 + H)+ ion intensities (Table 4) The highest R(MS3) among all the combinations of target and photoactive peptides was obtained when L* was near the C-terminus. The combined yields, which were calculated as R(total) = R(MS2) × R(MS3), were in the range of 21–30% (Table 2).
Sequence Analysis of Proline-Containing Cross-Links
The CID-MS3 spectra showed a number of sequence fragment ions originating from dissociations within the target (m) and denitrogenated photopeptide (M) chains. These were used for assigning the cross-link sites in the target peptides, following the previously reported procedure [10, 13]. Briefly, several criteria and features were used in the analysis. First, only the diazirine-bearing L* residue can be involved in photocross-linking, and thus, all logical backbone fragments must contain it. For example, fragment ions that would indicate cross-links at the C-terminal Y1, Y2, and Y3 sequences would be illogical for GL*LLK photopeptides and therefore would have been excluded from the analysis. Second, only fragment ions resulting from backbone cleavage in the target peptide can be used for cross-link assignment. The total relative intensities of sequence-specific fragment ions of the ymM, bnM, ym,Bn, and ymYq type varied for the different combinations of the target and photo-labeled peptides. It should be noted that when the target peptide contained proline, backbone cleavages upon CID were not likely to occur uniformly at all amide bonds because of the well-known proline effect, enhancing dissociation of the CO-N(Pro) amide bond [42,43,44,45]. It is also difficult to assess how a cross-link at a given residue affects backbone dissociations of the target peptide. However, the charging proton position in the complexes was firmly established in the photopeptide and its denitrogenated photoproduct, as revealed by the CID and UVPD mass spectra (Fig. 2b, c). It is also of note that there were a few isobaric combinations of the Pro and (L* – N2) (both 97 Da) and Leu residues that were then assigned to both possible chain combinations. These factors may result in over counting sequential C-terminal and N-terminal cross-links when based on ynM* and bnM* fragment ions, respectively. With all these caveats considered, the results of sequence analysis of all 15 Pro-containing complexes are displayed in Fig. 6a–e. The fractions of cross-links were normalized to the overall cross-linking yield for each photopeptide-target-pair.
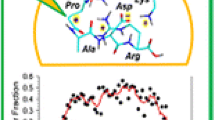
Cross-link distributions in proline target peptides. (a) PGLMG, (b) GPLMG, (c) GLPMG, (d) GMLPG, (e) GLMGP. The fractions were normalized to the R(total) efficiencies for each photopeptide-target peptide pair (cf. Table 2)
The data demonstrate that the Pro position in the target peptide had a substantial effect on the specificity of the carbene insertion and cross-link formation (Fig. 6a–e). Differences were also observed for cross-linking of the same target peptide with photopeptides having the L* residue in different positions. Starting with PGLMG, the CID-MS3 data indicated a tendency for cross-links with GL*LLK and GLLL*K to increasingly occur at residues close to the C-terminus of this target peptide (Fig. 6a). In contrast, cross-links to GLL*LK were more evenly distributed among the target peptide residues, with largest fractions appearing at Leu and Met. Analysis of the CID-MS2 spectrum of the target peptide ion alone, (PGLMG + H)+ (Fig. S37a, Supporting Information), indicated enhanced cleavage of the Met4-Gly5 and Leu3-Met4 amide bonds. This indicated that cross-links at Met4 and Gly5 could be somewhat overestimated in the sequence analysis of the complexes. A relatively non-selective distribution of cross-links among the amino acid residues was obtained for photodissociation of GPLMG with all three photopeptides (Fig. 6b). The target peptide ion, (GPLMG + H)+, was found to favor cleavage of the Met4-Gly5 and Leu3-Met4 amide bonds (Fig. S37b, Supporting Information), indicating again that cross-links at Met4 and Gly5 in the complexes could be somewhat overestimated by the sequence analysis.
The CID-MS3 data for GLPMG indicated highly preferential cross-linking at the Pro3, Met4, and C-terminal Gly5 residues (Fig. 6c). The fragment ion intensities are affected by the facile backbone cleavage at the Pro3 residue, leading to the dominant y3M ion at m/z 830 (Fig. 3). Note, however, that the sequence-complementary b2(M,Y,B) fragment ions were much less intense than the y3M ion, indicating less efficient cross-linking within the N-terminal Gly1-Leu2 sequence segment. The distinction of cross-links at Pro3, Met4, and C-terminal Gly5 was more difficult to achieve. The reference CID-MS2 spectrum of the target peptide ion, (GLPMG + H)+ (Fig. S37c, Supporting Information), showed a dominant dissociation of the Leu2-Pro3 amide bond, and thus, cross-links at Pro3, Met4, and Gly5 were expected to give rise to y3M ions regardless of the specific cross-link position within the PMG segment. However, specific cross-linking at C-terminal Gly5 and Met4 was indicated by the respective y1M (m/z 602) and y2M (m/z 733) fragment ions (Fig. 3). An interesting feature of the CID-MS3 data for GLPMG was the very similar cross-link distributions originating from GL*LLK, GLL*LK, and GLLL*K, and indicating preferential interactions with the target peptide C-terminal residues for all three photopeptides.
The proline effect on backbone dissociations also played a role in affecting the CID-MS3 data for GMLPG (Fig. 6d). The reference CID-MS2 spectrum of (GMLPG + H)+ exhibited a prominent y2 fragment ion by Leu3-Pro4 amide bond cleavage (Fig. S37d, Supporting Information) which could hamper distinction of cross-links at Pro4 and Gly5 in the complexes. However, regardless of the L* residue position in the photopeptide, the data indicated preferential interactions and cross-linking at the C-terminal Pro4 and Gly5 residues. Cross-links at Pro4 and C-terminal Gly5 were indicated by the respective y2M and y1M fragment ions in the CID-MS3 spectrum (Fig. S23–S25). Again, the sequence-complementary b3(M,Y,B) fragment ions were much less intense than the y2M ion, indicating less efficient cross-linking at the N-terminal Gly1, Met2, and Leu3 residues of the target peptide.
Cross-linking of the Pro-C-terminal target peptide GLMGP displayed dependence on the position of the L* residue (Fig. 6e). With GL*LLK and GLL*LK, a broad distribution of cross-links was formed that peaked around the middle Met3 residue. In contrast, with GLLL*K, the majority of cross-links were formed at the Leu2, Met3, and N-terminal Gly1 residues. These results indicated different interactions with the target peptide of the carbene intermediates when generated at different positions of the photopeptide. We note that the reference CID-MS2 spectrum of the target peptide ion (GLMGP + H)+ indicated preferred dissociation at Gly4-Pro5, and hence, the assignment of cross-links at the N-terminal residues in the complex would not be seriously affected by a similar dissociation bias.
Phenylalanine-Containing Peptides
Photocross-linking of the Phe-containing peptides markedly differed from that of the Pro-containing peptides, as is evident from the comparison of Figs. 6 and 7 distributions. In particular, as shown in Fig. 7, the neutral peptide GFLMG did not prefer a particular photocross-linking position with any photopeptide counterparts. However, as the Phe and L* residues moved towards the C-termini of their respective sequences, the cross-linking yield increased. In the case of GMLFG, the most efficient cross-linking was at the C-terminal Gly5 in a complex with GLLL*K. Similarly, GLFMG showed prominent cross-linking at Gly5 when interacting with GLLL*K. The increase in the yield of photocross-linking among Phe-containing peptides may be accounted for by the interaction between the π-system of the Phe ring and the charge at the Lys ε-amine in the photopeptide. This π-system interaction would favor close positioning of the L* and Phe residues, subsequently increasing the probability of cross-linking at positions nearby phenylalanine.
Complex Ion Structures and Born-Oppenheimer Molecular Dynamic Analysis
In order to further elucidate the non-covalent interactions of Pro-containing complexes, we selected the (GLPMG + GLL*LK + H)+ complex as our model system for BOMD trajectory calculations. One of the reasons for selecting this peptide pair over the others was the middle position of the proline residue within the target peptide sequence. BOMD analysis could potentially resolve the ambiguity in determining the position of cross-links within the Pro-Met-Gly sequence (vide supra) and provide structures of the complexes. The computational analysis was performed in three steps. In the first step, we determined the structures of lowest free-energy complexes as local minima at 0 K. In the next step, we worked out the thermodynamics of the complexes to establish the equilibrium constants and mole fractions in the gas phase. In the last step, we used the energy-sorted complexes for long (100 ps) BOMD trajectory calculations.
The relative free energies of (GLPMG + GLL*LK + H)+ complexes, (ΔG310, Table S1), were based on ωB97X-D energies for fully optimized structures, whereas enthalpies and entropies were taken from B3LYP frequency calculations. The ωB97X-D functional was relied on because it includes dispersion interactions that we deemed to be important in non-covalent complexes. The free energies were then used to calculate pair-wise equilibrium constants Kin and mole fractions xin, Kin = exp(−ΔGin/RT), xi = Kin/∑Kin where n refers to the reference complex and i = 1,..,12 (Table S2). The calculations identified five isomers (1–5) that had ΔG310 within 25 kJ mol−1 of the lowest energy complex 2, from which we obtained the expected mole fractions of 0.80, 0.19, 0.008, 0.0004, and 0.00008 for 2, 1, 5, 3, and 4, respectively. Structure-wise, complexes 2–5 had canonical structures with a protonated Lys side chain in GLL*LK and a neutral target peptide (Fig. 8). This is consistent with the CID-MS2 experiments that showed prevalent proton retention in the photopeptides (Fig. 2b). Structure 1 of a slightly higher ΔG310 than 2 was a complex of a GLL*LK zwitterion with GLPMG protonated at the N-terminus (Fig. 8). Complexes 6–12 had higher ΔG310 excluding them from being substantially populated in an equilibrium mixture (Table S2). Out of these, complexes 7, 8, 9, 11, and 12 were canonical structures, whereas 6 and 10 were zwitterions (Fig. S38, Supporting Information). The non-covalent bonding in all these isomers was mediated by hydrogen bonds of the charged NH3 and neutral COOH groups to amide electron donors in the peptide counterpart. For example, complex 1 displayed four strong intermolecular hydrogen bonds that were between the Lys-5 carboxylate and Gly5 carboxyl, and Leu4-Gly5, Leu4-Pro3, and Leu2-Leu2 amides (Fig. 8). In the lowest free-energy complex 2, there were strong intermolecular H-bonds between Gly1-Gly5, Lys5-NH3+-Pro3, and Lys5-Leu2. The third most stable complex 5 displayed H-bonding between Lys5-NH3+, Leu4, and the Gly5 carboxyl group.
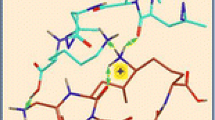
Left panel: ωB97X-D/6-31 + G(d,p) optimized structures of low free-energy (GLPMG + GLL*LK + H)+ complexes 1–5. Green arrows indicate major hydrogen bonds between the photopeptide and target peptide. Asterisks indicate the diazirine rings. Right panel: Target peptide geometries in the complexes. Green arrows indicate major hydrogen bonds within the target peptide. Atom color coding: Cyan or magenta = C, gray = H, blue = N, red = O, yellow = S. Only exchangeable (N–H, O–H) hydrogen atoms are shown
The hydrogen bonding pattern had a significant effect on the target peptide secondary structure. This is shown in the right hand panel of Fig. 8 depicting the target GLPMG peptide moieties in conformations with which they appear in the complexes. The structures reveal that the target peptides in 1, 3, 4, and 5 folded to form β turns at Pro. The hairpin conformations were cooperatively favored by intramolecular H-bonding of the Pro-flanking residues in the target peptides, and intermolecular H-bonding to the photopeptide. The lowest free-energy complex 2 was exceptional in that the target peptide unit was extended by intermolecular H bonding (vide supra) and did not form a β turn at Pro.
Long (100 ps) BOMD trajectories were run at 310 K and analyzed for low-energy (GLPMG + GLL*LK + H)+ conformers 1–12. The analysis yielded close contacts between the diazirine carbon (C35) on L* and X–H bonds in the target peptide. Close contacts were identified for all C35—X distances within 4.5 Å, corresponding to a projection of a sum of van der Waals radii of atoms constituting the diazirine ring and X–H bonds [10]. The numbering system for the close contact analysis is depicted in Table S3 (Supporting Information). The BOMD analysis provided insights regarding the intermolecular and intramolecular interactions between the peptide components in the complexes under thermal motion. This is illustrated by the close contact data in Table S3, which shows the percentage of time during the 100 ps BOMD run, consisting of 100,000 steps, when the carbon atom (C35) of the diazirine ring was within 4.5 Å of the hydrogen carrying atoms of the target peptide. For example, in complex 1, C35 was within the contact distance with the C100 methylene of the Leu2 residue in the target peptide for 4.4% of the time, corresponding to 4429 contacts, while not getting into contact with the X–H atoms on the other residues of the target peptide. Regarding the lowest-energy complex 2, BOMD showed multiple (153353) C35 contacts that were chiefly with atoms at the Met4 and Gly5 residues of the target peptide. However, the fraction of contacts with the Gly5 COOH carbonyl group were discounted because they could not result in an X–H bond insertion, thus reducing the number of potentially reactive contacts to 132,304.
The BOMD analysis of close contacts in (GLPMG + GLL*LK + H)+ in 1–12 (Table S3) was combined with the calculated populations of these conformational isomers (Table S2) to estimate the overall potential cross-linking sites in complexes existing at thermal equilibrium. When based on molar fractions obtained from the ωB97X-D free energies, the population-averaged cross-link distribution was 0.0, 1.0, 1.1, 97, and 0.7% at Gly1, Leu2, Pro3, Met4, and Gly5, respectively. An evaluation using B3LYP free energies and molar fractions gave a similar population-averaged breakdown of 0, 0.1, 0.2, 79, and 21% for cross-links at Gly1, Leu2, Pro3, Met4, and Gly5, respectively.
This result is consistent with the experimental distribution of cross-links obtained from gas-phase sequencing (Fig. 6) where the majority (> 90%) of the observed cross-links were assigned to the Pro3-Met4-Gly5 segment. As discussed above, distinction within this segment was difficult because of the proline effect favoring CID at the N-terminal side of Pro. Thus upon CID, complexes cross-linked at Pro3, Met4, and Gly5 can be expected to undergo abundant backbone cleavage at Pro3 to give the dominant y3M fragment ions without distinguishing the cross-link position within the Pro3, Met4, and Gly5 residues. In contrast, the proline effect did not affect the identification of cross-links at Gly1 and Leu2 because the complementary b2M fragment ions should be readily formed by backbone cleavage at Pro3. The experimental difficulty with CID backbone cleavage was resolved by the BOMD contact analysis. This indicated that Met4 and Gly5 were the most frequently visited residues, whereas Pro3 was not likely (1%) to be involved in cross-linking. We conclude that, in accord with experiment, the majority of cross-links in the (GLPMG + GLL*LK + H)+ complex were formed in the Met4 side chain of the lowest-free energy conformer 2.
The importance of BOMD analysis of contacts due to thermal motion in the complexes (Table S3) was made evident by comparison to static contacts in geometries of the local energy minima of 1–5 (Fig. 8), corresponding to 0 K structures. Complex 1 showed no close contacts between the incipient carbene (C35) and the target peptide residues in the 0 K structure. The X–H atom in the target peptide which was closest to C35 was C115 of the Pro residue at 6.14 Å. However, thermal motion in the complex did not result in a close contact with Pro X–H atoms. Rather, C35 developed close contacts with the flexible Leu2 side chain as a result of thermal motion. Similar conclusions followed from analysis of 0 K structures and thermally induced close contacts in all the other complexes. The majority of close contacts occurred in regions that were close to C35 in 0 K structures or could be approached via rotations of the Leu2 and Met4 side chains or the N-terminal Gly. These results strongly indicated that the hydrogen bonding framework of the complexes was not disrupted by thermal motion at 310 K. Our previous analysis of conformational changes in monomeric peptide ions indicated that hydrogen bond rearrangements occurred via a slipping motion, whereby the energy needed to disrupt one hydrogen bond was compensated by cooperative formation of another hydrogen bond [46,47,48]. The present results for peptide-peptide ion complexes indicated no such slippage, as the main hydrogen bonds connecting the peptide moieties were not rearranged by thermal motion. This can be attributed to a stable core framework of polar groups and hydrogen bonds that resist substantial rearrangement in thermal complexes.
Conclusions
The combination of diazirine photocross-linking, ion activation, tandem mass spectrometry (UVPD/CID), and all-valence-electron molecular dynamics allowed us to gain insight regarding the non-covalent interactions of neutral proline- and phenylalanine-containing peptides in gas-phase complexes. The structures of the gas-phase complexes were maintained by a stable core framework of polar groups and hydrogen bonds that did not substantially rearrange by thermal motion. Polar non-covalent bonding in the complexes had a substantial effect on the secondary structure of the target peptide. Close contacts in the complexes leading to cross-links in carbene intermediates predominantly occurred in residues that were suitably positioned already in the 0 K structures. The experimental data indicated that diazirine photodissociation resulted in 19–37% fractions of covalent cross-links. We conclude that gas-phase cross-linking using diazirine-tagged peptides represents a high-yield method that furthermore provides experimental background for detailed structural information at atomic-level resolution to be obtained by valence-electron molecular dynamics calculations. Applications of this combined approach to elucidations of non-covalent peptide-peptide interactions are in progress in this laboratory.
References
Fancy, D.A., Kodadek, T.: Chemistry for the analysis of protein–protein interactions: rapid and efficient cross-linking triggered by long wavelength light. Proc. Natl. Acad. Sci. U. S. A. 96, 6020–6024 (1999)
Das, J.: Aliphatic diazirines as photoaffinity probes for proteins: recent developments. Chem. Rev. 111, 4405–4417 (2011)
Singh, A., Thornton, E.R., Westheimer, F.H.: Photolysis of diazoacetylchymotrypsin. J. Biol. Chem. 237, PC3006–PC3008 (1962)
Fleet, G.W.J., Porter, R.R., Knowles, J.R.: Affinity labeling of antibodies with aryl nitrene as reactive group. Nature (London). 224, 511–512 (1969)
Knowles, J.R.: Photogenerated reagents for biological receptor-site labeling. Acc. Chem. Res. 5, 155–160 (1972)
Liu, M.T.H.: Chemistry of diazirines, vol. I and II. CRC Press, Boca Raton (1987)
Tomioka, H., Izawa, Y.: Photolysis of aryldiazomethanes in alcoholic matrices. Temperature and host dependencies of phenylcarbene processes in alcohols. J. Am. Chem. Soc. 99, 6128–1629 (1977)
Senthilnathan, V.P., Platz, M.S.: Determination of the absolute rates of decays of arylcarbenes in various low temperature matrices by electron spin resonance spectroscopy. J. Am. Chem. Soc. 102, 7637–7643 (1980)
Dix, E.J., Goodman, J.L.: Protonation of diarylcarbenes by alcohols: the importance of ion pair dynamics. J. Phys. Chem. 98, 12609–12612 (1994)
Shaffer, C.J., Andrikopoulos, P.C., Řezáč, J., Rulíšek, L., Tureček, F.: Efficient covalent bond formation in gas-phase peptide–peptide ion complexes with the photoleucine stapler. J. Am. Soc. Mass Spectrom. 27, 633–645 (2016)
Suchanek, M., Radzikowska, A., Thiele, C.: Photo-leucine and photo-methionine allow identification of protein-protein interactions in living cells. Nat. Methods. 2, 261–268 (2005)
Yang, T., Li, X.-M., Bao, X., Fung, Y.M.E., Li, D.: Photolysine captures proteins that bind lysine post-traslation modifications. Nat. Chem. Biol. 12, 70 (2016)
Pepin, R., Shaffer, C.J., Turecek, F.: Position-tunable diazirine tags for peptide-peptide ion cross-linking in the gas phase. J. Mass Spectrom. 52, 557–560 (2017)
Sinz, A., Arlt, C., Chorev, D., Sharon, M.: Chemical cross-linking and native mass spectrometry: a fruitful combination for structural biology. Protein Sci. 24, 1193 (2015)
Lee, S., Valentine, S.J., Reilly, J.P., Clemmer, D.E.: Controlled formation of peptide bonds in the gas phase. J. Am. Chem. Soc. 133, 15834–15837 (2011)
McGee, W.M., McLuckey, S.A.: Efficient and directed peptide bond formation in the gas phase via ion/ion reactions. Proc. Natl. Acad. Sci. U. S. A. 111, 1288–1292 (2014)
Koelbel, K., Warnke, S., Seo, J., von Helden, G., Moretti, R., Meiler, J., Pagel, K., Sinz, A.: Conformational shift of a β-hairpin peptide upon complex formation with an oligo-proline peptide studied by mass spectrometry. Chem. Select. 1, 3651 (2016)
Pezacki, J.P., Couture, P., Dunn, J.A., Warkentin, J., Wood, P.D., Lusztyk, J., Ford, F., Platz, M.S.: Rate constants for 1,2-hydrogen migration in cyclohexylidene and in substituted cyclohexylidenes. J. Org. Chem. 64, 4456 (1999)
Schimmel, P.R., Flory, P.J.: Conformational energies and configurational statistics of copolypeptides containing L-proline. J. Mol. Biol. 34, 105–120 (1968)
Lewis, P.N., Momany, F.A., Scheraga, H.A.: Folding of polypeptide chains in proteins: a proposed mechanism for folding. Proc Nat. Acad. Sci. U.S.A. 68, 2293–2297 (1971)
Kuntz, I.D.: Protein folding. J. Am. Chem. Soc. 94, 4009–4012 (1972)
Crawford, J.L., Lipscomb, W.N., Schellman, C.G.: The reverse turn as a polypeptide conformation in globular proteins. Proc. Natl. Acad. Sci. U. S. A. 70, 538–542 (1973)
MacArthur, M.W., Thornton, J.M.: Influence of proline residues on protein conformation. J. Mol. Biol. 218, 397–412 (1991)
Shi, L., Holliday, A.E., Bohrer, B.C., Kim, D., Servage, K.A., Russell, D.H., Clemmer, D.E.: Wet versus dry folding of polyproline. J. Am. Soc. Mass Spectrom. 27, 1037–1047 (2016)
Basic Local Alignment Search Tool (BLAST) Available at: http://blast.ncbi.nlm.gov. Accessed 10 June 2016
Řezáč, J., Fanfrlík, J., Salahub, D., Hobza, P.: Semiempirical quantum chemical PM6 method augmented by dispersion and H-bonding correction terms reliably describes various types of noncovalent complexes. J. Chem. Theor. Comput. 5, 1749–1760 (2009)
Janz, J.M., Ren, Y., Looby, R., Kazmi, M.A., Sachdev, P., Grunbeck, A., Haggis, L., Chinnapen, D., Lin, A.Y., Seibert, C., McMurry, T., Carlson, K.E., Muir, T.W., Hunt, S., Sakmar, T.P.: Direct interaction between an allosteric agonist pepducin and the chemokine receptor CXCR4. J. Am. Chem. Soc. 133, 15878–15881 (2011)
Coste, J., LeNguyen, D., Castro, B.: PyBOP: a new peptide coupling reagent devoid of toxic by-product. Tetrahedron Lett. 31, 205–208 (1990)
Shaffer, C.J., Marek, A., Nguyen, H.T.H., Tureček, F.: Combining near-UV photodissociation with electron transfer. Reduction of the diazirine ring in a photomethionine-labeled peptide ion. J. Am. Soc. Mass Spectrom. 26, 1367–1381 (2015)
Berendsen, H.J., Postma, J.V., van Gunsteren, W.F., DiNola, A.R.H.J., Haak, J.R.: Molecular dynamics with coupling to an external bath. J. Chem. Phys. 81, 3684–3690 (1984)
Řezáč, J.: Cuby—ruby framework for computational chemistry, version 4, http://cuby4.molecular.cz Accessed June 2016.
Řezáč, J.: Cuby: an integrative framework for computational chemistry. J. Comput. Chem. 37, 1230–1237 (2016)
Becke, A.D.: Density-functional exchange-energy approximation with correct asymptotic behavior. Phys. Rev. A. 38, 3098–3100 (1988)
Lee, C., Yang, W., Parr, R.G.: Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys. Rev. B37, 785–789 (1988)
Schäfer, A., Horn, H., Ahlrichs, R.: Fully optimized contracted Gaussian basis sets for atoms lithium to krypton. J. Chem. Phys. 97, 2571–2577 (1992)
TURBOMOLE V6.6. A development of University of Karlsruhe and Forschungszentrum Karlsruhe GmbH, 1989–2014, TURBOMOLE GmbH, since 2007, available at: http://www.turbomole.com 2014. Accessed 7 June 2017
Frisch, M.J., Trucks, G.W., Schlegel, H.B., Scuseria, G.E., Robb, M.A., Cheeseman, J.R., Scalmani, G., Barone, V., Mennucci, B., Petersson, G.A., Nakatsuji, H., Caricato, M., Li, X., Hratchian, H.P., Izmaylov, A.F., Bloino, J., Zheng, G., Sonnenberg, J.L., Hada, M., Ehara, M., Toyota, K., Fukuda, R., Hasegawa, J., Ishida, M., Nakajima, T., Honda, Y., Kitao, O., Nakai, H., Vreven, T., Montgomery Jr., J.A., Peralta, J.E., Ogliaro, F., Bearpark, M., Heyd, J.J., Brothers, E., Kudin, K.N., Staroverov, V.N., Kobayashi, R., Normand, J., Raghavachari, K., Rendell, A., Burant, J.C., Iyengar, S.S., Tomasi, J., Cossi, M., Rega, N., Millam, J.M., Klene, M., Knox, J.E., Cross, J.B., Bakken, V., Adamo, C., Jaramillo, J., Gomperts, R., Stratmann, R.E., Yazyev, O., Austin, A.J., Cammi, R., Pomelli, C., Ochterski, J.W., Martin, R.L., Morokuma, K., Zakrzewski, V.G., Voth, G.A., Salvador, P., Dannenberg, J.J., Dapprich, S., Daniels, A.D., Farkas, O., Foresman, J.B., Ortiz, J.V., Cioslowski, J., Fox, D.J.: Gaussian 09, revision A02. Gaussian, Inc., Wallingford CT (2009)
Frisch, M.J., Trucks, G.W., Schlegel, H.B., Scuseria, G.E., Robb, M.A., Cheeseman, J.R., Scalmani, G., Barone, V., Petersson, G.A., Nakatsuji, H., Caricato, M., Marenich, A.V., Bloino, J., Janesko, B.G., Gomperts, R., Mennucci, B., Hratchian, H.P., Ortiz, J.V., Izmaylov, A.F., Sonnenberg, J.L., Williams-Young, D., Ding, F., Lipparini, F., Egidi, F., Goings, J., Peng, B., Petrone, A., Henderson, T., Ranasinghe, D., Zakrzewski, V.G., Gao, J., Rega, N., Zheng, G., Liang, W., Hada, M., Ehara, M., Toyota, K., Fukuda, R., Hasegawa, J., Ishida, M., Nakajima, T., Honda, Y., Kitao, O., Nakai, H., Vreven, T., Throssell, K., Montgomery Jr., J.A., Peralta, J.E., Ogliaro, F., Bearpark, M.J., Heyd, J.J., Brothers, E.N., Kudin, K.N., Staroverov, V.N., Keith, T.A., Kobayashi, R., Normand, J., Raghavachari, K., Rendell, A.P., Burant, J.C., Iyengar, S.S., Tomasi, J., Cossi, M., Millam, J.M., Klene, M., Adamo, C., Cammi, R., Ochterski, J.W., Martin, R.L., Morokuma, K., Farkas, O., Foresman, J.B., Fox, D.J.: Gaussian 16, revision A01. Gaussian, Inc., Wallingford CT (2016)
Chai, J.D., Head-Gordon, M.: Systematic optimization of long-range corrected hybrid density functionals. J. Chem. Phys. 128, 084106/1–084106/15 (2008)
Chai, J.D., Head-Gordon, M.: Long-range corrected hybrid density functionals with damped atom-atom dispersion corrections. Phys. Chem. Chem.Phys. 10, 6615–6620 (2008)
Marek, A., Tureček, F.: Collision-induced dissociation of diazirine-labeled peptide ions. Evidence for Brønsted-acid assisted elimination of nitrogen. J. Am. Soc. Mass Spectrom. 25, 778–789 (2014)
Schwartz, B.L., Bursey, M.M.: Some proline substituent effects in the tandem mass spectrum of protonated penta-alanine. Biol. Mass Spectrom. 21, 92–96 (1992)
Vaisar, T., Urban, J.: Probing the proline effects in CID of protonated peptides. J. Mass Spectrom. 31, 1185–1187 (1996)
Loo, J.A., Edmonds, C.G., Smith, R.D.: Tandem mass spectrometry of very large molecules. 2. Dissociation of multiply charged proline-containing proteins from electrospray ionization. Anal. Chem. 65, 425–438 (1993)
Savitski, M.M., Kjeldsen, F., Nielsen, M.L., Zubarev, R.A.: Complementary sequence preferences of electron-capture dissociation and vibrational excitation in fragmentation of polypeptide polycations. Angew. Chem. Int. Ed. 45, 5301 (2006)
Pepin, R., Laszlo, K.J., Peng, B., Marek, A., Bush, M.F., Tureček, F.: Comprehensive analysis of Gly-Leu-Gly-Gly-Lys peptide dication structures and cation-radical dissociations following electron transfer: from electron attachment to backbone cleavage, ion-molecule complexes and fragment separation. J. Phys. Chem. A. 118, 308–324 (2014)
Tureček, F., Chen, X., Hao, C.: Where does the electron go? Electron distribution and reactivity of peptide cation-radicals formed by electron transfer in the gas phase. J. Am. Chem. Soc. 130, 8818–8833 (2008)
Nguyen, H.T.H., Andrikopoulos, P.C., Bím, D., Rulíšek, L., Dang, A., Tureček, F.: Radical reactions affecting polar groups in threonine peptide ions. J. Phys. Chem. B. 121, 6557–6569 (2017)
Funding
Research at University of Washington has received support from the Chemistry Division of the National Science Foundation (Grants CHE-1543805, CHE-1661815, and CHE-1624430). F.T. thanks the Klaus and Mary Ann Saegebarth Endowment for general support. Research at the IOCB Prague was supported by the Grant Agency of the Czech Republic (grant 17-24155S).
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
ESM 1
(PDF 2576 kb)
Rights and permissions
About this article
Cite this article
Nguyen, H.T.H., Andrikopoulos, P.C., Rulíšek, L. et al. Photodissociative Cross-Linking of Non-covalent Peptide-Peptide Ion Complexes in the Gas Phase. J. Am. Soc. Mass Spectrom. 29, 1706–1720 (2018). https://doi.org/10.1007/s13361-018-1980-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13361-018-1980-4