Abstract
Matrix-assisted laser desorption ionization (MALDI) imaging mass spectrometry has been adopted in the pharmaceutical industry as a useful tool to detect xenobiotic distribution within tissues. A unique sample preparation approach for MALDI imaging has been described here for the extraction and detection of cobimetinib and clozapine, which were previously undetectable in mouse and rat brain using a single matrix application step. Employing a combination of a buffer wash and a cyclohexane pre-extraction step prior to standard matrix application, the xenobiotics were successfully extracted and detected with an 8 to 20-fold gain in sensitivity. This alternative approach for sample preparation could serve as an advantageous option when encountering difficult to detect analytes.

ᅟ
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Tissue imaging via matrix assisted laser desorption/ionization mass spectrometry (MALDI MS) is a common technique utilized in the pharmaceutical industry to detect the distribution of label-free drug within thin tissue sections [1–5]. In current practice, most analytes can be successfully extracted from tissue using a standardized sample preparation protocol where fresh frozen tissues, ranging from 10 to 20 μm thickness, are thaw-mounted onto a MALDI target plate and coated with a universal matrix solution. Matrix application is an integral step in the sample preparation process and serves two purposes. First, the solvent in which the matrix is dissolved helps to extract the analytes of interest onto the tissue surface. Then, as the solvent evaporates, the matrix co-crystalizes with the extracted analytes. The matrix co-crystals are necessary to assist in the desorption and ionization of nonvolatile analytes by facilitating the absorption of the laser energy and in turn transferring this energy to the analytes to produce ions in the gas phase. It is important to select the correct matrix solvent combination to ensure optimal extraction and ionization of analytes of interest. Generally, in order to detect peptides, proteins, lipids, and drugs, the three most commonly used matrices are 2,5-dihydroxybenzoic acid (DHB), α-cyano-4-hydroxycinnamic acid (CHCA), and 3,5-dimethoxy-4-hydroxycinnamic acid (SA, sinapinic acid) dissolved in 50%–70% organic solvent such as methanol, ethanol, or acetonitrile. Matrix concentration and organic solvent percentage, however, can be further tailored to the particular analyte of interest for increased extraction efficiency [6].
When matrix and solvent optimization is not enough, additional steps such as buffer washes are necessary to enhance analyte signal. Reported washes include aqueous buffer solutions like ammonium formate and ammonium acetate to enhance lipid signal in negative mode [7] and pH controlled ammonium acetate for small molecule drug signal enhancement [8]. Once washes are carried out, matrix can then be applied in various ways, which include manual spray coating, sublimation, microspotting, or dry-coating [9–13]. Each matrix application approach can have its own benefits and drawbacks.
Manual spray coating via thin-liquid chromatography (TLC) sprayer in the hands of a well-trained individual can yield optimal results since the operator can visually decide if the matrix coating is “wet” enough to extract the analytes but not “too wet” to cause delocalization [12]. In turn, spraying manually can be cumbersome as it must be applied with great skill in order to produce consistent, homogenous coverage of the tissue. Sublimation and dry-coating are both solvent-free coating techniques that eliminate the possibility of delocalization, but its application has been limited to lipids and highly abundant species at the tissue surface since minimal extraction is being performed from the tissue because of the lack of solvent [10, 11]. Microspotting is a technique that involves the deposition of matrix via a piezoelectric device that prints an array of matrix microspots (~pL-nL volumes) at the tissue surface [9, 13]. These systems allow for the complete control of matrix placement on the tissue, and prolonged solvent extraction as the larger droplets dry. However, image resolution is sacrificed as one can only image where matrix spots have been deposited and limits experiments to instruments with software that allow for spotted array acquisitions.
More recently, examples of matrix precoated MALDI target plates have been reported where plates are homogenously coated with matrix prior to the thaw-mounting of tissues [14, 15]. This method is beneficial for individuals looking for a fast and easy sample preparation method since tissues can be mounted onto the precoated plate and directly analyzed in the mass spectrometer without the need for further processing. However, this technique has only been demonstrated with the detection of highly abundant species since minimal extraction from the tissue is occurring. In the event tissue washing steps are needed, this technique cannot be utilized since the matrix precoat would wash away prior to analysis. For the experiments presented here, a robotic sprayer was utilized since previous examples have shown that robust and reproducible homogenous matrix coatings with little to no delocalization can be routinely achieved [16–18]. Drawbacks of utilizing a robotic sprayer, however, include the fact that there are many customizable options. Various parameters need to be optimized, such as flow rate, velocity, track spacing, nozzle spray temperature, and nozzle height, all of which require patience and diligence to identify the best combination. Further, these parameters have been shown to be dependent on the matrix and solvents used for spraying. Thus, in the interest of throughput, set universal sample preparations using the robotic sprayer are limited to one or two matrix solvent combinations.
Hence, the method of matrix application is an important consideration in the MALDI MS imaging process to ensure efficient extraction of analytes prior to detection in a mass spectrometer. Unfortunately, in some special cases, a single extraction step during the matrix application may not be sufficient to extract the analytes and may lead to a false conclusion that analytes are absent from the tissue, especially without any additional quantitative information. To address this challenge, additional method development was attempted to improve extraction of particularly challenging compounds encountered in the drug discovery setting where MALDI MS imaging is utilized to detect compound distribution. It was hypothesized that factors contributing to the lack of drug signal may include scenarios where compounds are highly bound to tissue/target and cannot be efficiently extracted by the solvents utilized in standard matrix solutions, such as acetonitrile and methanol. Therefore, additional solvents were explored to improve extraction efficiency during the sample preparation stage. Ultimately, an alternative sample preparation approach that combined both a wash and solvent pre-extraction step prior to matrix deposition was developed to enable the analysis of compounds that were previously undetectable under the standard preparation conditions.
Methods
Materials
2,5-Dihydroxybenzoic acid (DHB) matrix, clozapine, ammonium formate, ammonium citrate, cresyl violet and cyclohexane were purchased from Sigma Chemical Co. (St. Louis, MO, USA). Cobimetinib (GDC-0973), 13C6-cobimetinib and a proprietary compound (Genentech Compound X) used as an internal standard were synthesized by Genentech Inc. (South San Francisco, CA, USA). Methanol was purchased from EMD Millipore Corporation (Billerica, MA, USA).
Tissue Preparation
Mdr1a/1b/Bcrp1(−/−) Taconic mice were administered a single 10 mg/kg oral dose of cobimetinib. Sprague-Dawley rats were administered a single 8 mg/kg oral dose of clozapine. Mice at 2 h post-dose and rats at 6 h post-dose were euthanized via exsanguination by perfusion with PBS under anesthesia. Brains were excised, flash-frozen in liquid N2 and stored in a –80°C freezer until analyzed. Serial sections from the same subject were collected on a cryomicrotome (CM3050S; Leica, Buffalo Grove, IL, USA) at 12-μm thickness and thaw-mounted onto indium tin oxide-coated glass slides (Bruker Daltonics, Billerica, MA, USA).
Analyte Pre-Extraction and Preparation for Imaging
All mouse and rat serial sections were thaw-mounted on the same glass slide, yielding two slides total, one for mouse brains and one for rat brains. A representative section was selected for each of the various treatment scenarios. This was repeated three times. Tissue sections were washed three times at 5 s intervals with either 50 mM ammonium citrate or 50 mM ammonium formate. After drying, sections were homogenously precoated with cyclohexane using the automated HTX TM-Sprayer (HTX Technologies, Chapel Hill, NC, USA). Approximately 6 mL of cyclohexane were sprayed onto the tissue section with a nozzle temperature of 25°C and a solvent flow rate of 0.3 mL/min. A total of eight passes with a dry time of 0.5 min in between passes were carried out with 3 mm line spacing and a spray nozzle velocity of 1200 mm/min. This extraction step was immediately followed by a spray of 2,5-dihydroxybenzoic acid (DHB) (Sigma-Aldrich) matrix (40 mg/mL, 70/30 methanol/H2O) with a 200 nM internal standard spike of 13C6-cobimetinib for brains dosed with cobimetinib and 2 uM internal standard spike of Genentech Compound X for brains dosed with clozapine. Matrix was applied utilizing the same HTX TM-Sprayer method used to spray cyclohexane with a modified nozzle temperature of 75°C and solvent flow rate of 0.2 mL/min.
Imaging MALDI MS
Each slide was coated with matrix at the same time following the various wash and solvent spray pretreatments. Matrix-coated tissue sections were transferred and analyzed by imaging MALDI mass spectrometry (MS) on a 7.0 T SolariX FT-ICR system (Bruker Daltonics, Bremen, Germany) equipped with a dual ESI-MALDI source employing Smartbeam-II technology. Clozapine MS spectra were acquired in positive ion mode, in broadband with the mass range of m/z 100–3000 at 200 μm pixel resolution. Cobimetinib was acquired in positive ion mode utilizing continuous accumulation of selected ions (CASI) for a single accumulation window optimized for 5 Da centered on the parent mass of cobimetinib with ion detection from m/z 150–3000 and a pixel resolution of 150 μm. Laser intensity and the number of shots were optimized for each internal standard response (~700–1000 shots). For each mass spectrometry method used to detect clozapine and cobimetinib, a data reduction of 97% was applied with an internal standard lock mass of m/z 401.072117 and 538.090465, respectively. Regions to be imaged were drawn outside the tissue border in order to include several pixels representing non-tissue. Ion images were extracted based on the accurate mass of each compound (cobimetinib = m/z 532.07034; clozapine = m/z 327.13710), normalized to their internal standard response, and displayed using FlexImaging software v4.2 64-bit (Bruker Daltonics, Bremen, Germany) with a mass tolerance of ±5 mDa. Overall average intensity counts were determined by drawing regions of interests around the brain tissue pixels and exporting into Data Analysis v4.2 64-bit (Bruker Daltonics).
Histologic Staining
After imaging by MALDI MS, brain sections were stained by cresyl violet for histologic interrogation. To do so, the matrix coating was first rinsed away with 100% methanol, then stained with a freshly prepared 0.5% cresyl violet staining solution [19]. To stain, the glass slide was submerged in the cresyl violet staining solution for 30 s, and subsequently rinsed in 100% ethanol for two cycles at 30 s each. Microscope images were obtained on an Olympus BX51 (Olympus, Tokyo, Japan) at 10× magnification and stitched using MicroSuite Analytical v3.0 software (Olympus).
Results and Discussion
In MALDI MS imaging, most analytes of interest can be detected utilizing a standard sample preparation step, which involves the deposition of a MALDI matrix to facilitate the extraction and desorption of analytes. However, in some special cases where the analytes of interest are present but cannot be detected utilizing the standard single extraction step, different extraction methods should be explored. Previous work had demonstrated the utilization of ammonium formate buffer wash to enhance lipid analysis by negative ion mode and ammonium acetate to enhance the signal of targeted small molecules without spatial delocalization [7, 8]. Encouraged by these promising results, experiments were carried out to test the effectiveness of buffer washing combined with cyclohexane pre-extraction on brain tissues. Presented here is an alternative analyte pre-extraction approach that enables a path to detect previously undetectable small molecule compounds.
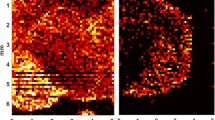
In the case of imaging cobimetinib from mouse brain, the standard matrix application of DHB in 70% methanol yielded barely detectable signal in tissue (Figure 1b). Based on quantitative data obtained from brain homogenates [1], it was concluded that levels of drug in brain were within the detectable range of the MALDI MS assay (sub-uM). This was confirmed by spotting a standard of equivalent concentration on a blank tissue section, spray coated, and imaged via MALDI MS with the limit of detection determined to be 210 nM/pixel. A representative spectrum from the spot demonstrates adequate sensitivity and signal to noise can be achieved by MALDI MS (Figure 2). It was then hypothesized that inadequate extraction of the drug during the matrix application process may be the reason for the poor MALDI sensitivity. Subsequently, with the quantitative knowledge in hand, cyclohexane was tested to assist in the extraction of drug prior to matrix application. This extraction pre-spray step with cyclohexane proved successful in enhancing cobimetinib signal (Figure 1c) without delocalization, as there was no signal detected beyond the tissue border. Comparing the signal intensity of cobimetinib from a standard DHB matrix alone with that of a tissue presprayed with cyclohexane, the overall average intensity counts of the drug doubled (Table 1). The corresponding cobimetinib ion image confirmed a more ubiquitous detection of drug signal across the brain, compared with the brain section that received matrix only (Figure 1c). However, a wash step was also incorporated to determine if the drug signal could be further enhanced. The use of an ammonium citrate wash was able to produce an ion image that could detect cobimetinib across the entire brain section, with enhanced signal localization in the cortex region and little to no signal in the white matter regions (Figure 1d). Comparing the overall average signal counts with matrix alone indicated that the ammonium citrate wash, prior to matrix application, was able to dramatically increase detection by ~17-fold (Table 1). When coupling the buffer wash and cyclohexane prespray, signal increased by ~20-fold (Table 1). Qualitative assessment of the ion image points to a more uniform detection of signal across the brain, with localization of cobimetinib also present in the white matter (Figure 1e). The overall average intensity counts of cobimetinib from the tissue images were plotted on the same spectrum to demonstrate the improvement in signal detection across the various sample preparation treatments (Figure 3). It is clear that the improvement in signal detection with ammonium citrate wash alone or in combination with cyclohexane now provides a wider dynamic range for detection across the tissue without the fear of delocalization.
Cobimetinib MALDI MS images collected at 150 um resolution. (a) Cresyl violet stained coronal mouse brain section. (b) Standard matrix application only. (c) Presprayed with cyclohexane, then with standard matrix application. (d) Washed with ammonium citrate, then standard matrix application. (e) Combined wash of ammonium citrate and a prespray of cyclohexane, followed by standard matrix application
Based on the encouraging results with cobimetinib, the novel sample preparation method was extended to rat brain sections dosed with clozapine, which demonstrated poor sensitivity in previous imaging studies (data not shown). Using the standard DHB matrix application step, clozapine could be detected from standard spots on control tissue with adequate sensitivity for the detection of isotopic peaks (Figure 4). Again, poor detection of clozapine was achieved from dosed tissue sprayed with DHB matrix alone (Figure 5b). Tissues were then presprayed with cyclohexane, which saw an improvement in ion detection (Figure 5c) and a ~3-fold gain in overall average signal intensity when compared to tissue sprayed with matrix only (Table 2). Next, tissues were treated with the identified ammonium citrate buffer wash, yielding an improvement in the ion image of clozapine (Figure 5d). The corresponding gain in overall average signal intensity was ~5-fold. As demonstrated with the previous cobimetinib experiments, tissue that was washed with ammonium citrate and presprayed with cyclohexane prior to matrix application yielded optimal extraction with a ~8-fold gain in overall average signal intensity (Table 2). As a result, a more ubiquitous distribution of clozapine was observed (Figure 5e) and is in line with previously reported autoradiography data, which detected clozapine distributed throughout the brain [20].
Clozapine MALDI MS images collected at 200 um resolution. (a) Cresyl violet stained sagittal rat brain section. (b) Standard matrix application only. (c) Presprayed with cyclohexane, then with standard matrix application. (d) Washed with ammonium citrate, then standard matrix application. (e) Washed with ammonium citrate and presprayed with cyclohexane, then standard matrix application
Ammonium formate, a more commonly used wash buffer [7, 8], was also tested to determine if it can yield the same signal gains as ammonium citrate. A quick experiment showed that tissue washed with ammonium formate also improved the ion image of clozapine (Supplemental Figure 1b) and increased overall average signal intensity by ~4-fold. It was interesting to note that compared with the ammonium citrate washes, the combination of ammonium formate wash coupled with the cyclohexane prespray behaved slightly better in signal gain (~9-fold overall average signal intensity gain) versus the ammonium citrate wash coupled with the cyclohexane prespray. The resulting ion image therefore displayed the same clozapine distribution but with slightly greater dynamic range of pixel intensities (Supplemental Figure 1c). The overall average intensity counts of clozapine from tissue images were plotted on the same spectrum to demonstrate the improvement in signal detection across the various sample preparation treatments (Figure 6). In the case of the clozapine, the ammonium formate wash yielded slightly better results than the ammonium citrate wash with the combination of wash and presprayed cyclohexane again giving the best results. Across all combinations tested in rat brain no analyte signal was detected beyond the tissue border. It was concluded that no delocalization of analyte occurred.
The MALDI ion images presented here demonstrate that each different pre-extraction step prior to matrix application can increase drug signal intensity. First, tissue was washed with ammonium citrate to wash away ionization-suppressing endogenous components, such as salts, lipids, and soluble interfering molecules, and then sprayed with cyclohexane, which acted as a pre-extraction solvent to bring analytes to the surface of the tissue prior to matrix application. Both steps on their own increase analyte sensitivity but when combined, the results yielded optimal sensitivity. This study demonstrated that additional method development can and should be carried out when initial studies reveal little to no detection of analyte signal, especially when accompanying analytical quantitation data indicate significant drug levels should be present in tissue. These studies bring to light that MALDI imaging results on their own should never be interpreted as definitive results; instead, they should be used as a complementary technique to evaluate drug distribution. MALDI images should be interpreted with caution as a lack of drug signal does not necessarily mean that drug is not present, especially in the absence of paired data such as LC-MS/MS and/or other imaging modalities (e.g., autoradiography). Further experiments will be necessary to determine if this technique is amenable to other tissues beyond the brain, as well as additional analytes with diverse physical-chemical properties. Additionally, this dataset demonstrated that pre-extraction techniques can be utilized without fear of displacing analytes of interest and is an encouraging technique to further explore for the imaging of drug distribution in tissues by MALDI MS.
References
Choo, E.F., Ly, J., Chan, J., Shahidi-Latham, S.K., Messick, K., Plise, E., Quiason, C.M., Yang, L.: Role of P-glycoprotein on the brain penetration and brain pharmacodynamic activity of the MEK inhibitor cobimetinib. Mol. Pharm. 11, 4199–4207 (2014)
Salphati, L., Shahidi-Latham, S., Quiason, C., Barck, K., Nishimura, M., Alicke, B., Pang, J., Carano, R.A., Olivero, A.G., Phillips, H.S.: Distribution of the phosphatidylinositol 3-kinase inhibitors Pictilisib (GDC-0941) and GNE-317 in U87 and GS2 intracranial glioblastoma models-assessment by matrix-assisted laser desorption ionization imaging. Drug Metab. Dispos. 42, 1110–1116 (2014)
Shahidi-Latham, S.K., Dutta, S.M., Prieto Conaway, M.C., Rudewicz, P.J.: Evaluation of an accurate mass approach for the simultaneous detection of drug and metabolite distributions via whole-body mass spectrometric imaging. Anal. Chem. 84, 7158–7165 (2012)
Prideaux, B., Dartois, V., Staab, D., Weiner, D.M., Goh, A., Via, L.E., Barry, C.E., 3rd, Stoeckli, M.: High-sensitivity MALDI-MRM-MS imaging of moxifloxacin distribution in tuberculosis-infected rabbit lungs and granulomatous lesions. Anal. Chem. 83, 2112–2118 (2011)
Castellino, S., Groseclose, M.R., Sigafoos, J., Wagner, D., de Serres, M., Polli, J.W., Romach, E., Myer, J., Hamilton, B.: Central nervous system disposition and metabolism of fosdevirine (GSK2248761), a non-nucleoside reverse transcriptase inhibitor: an LC-MS and matrix-assisted laser desorption/ionization imaging MS investigation into central nervous system toxicity. Chem. Res. Toxicol. 26, 241–251 (2013)
Norris, J.L., Caprioli, R.M.: Analysis of tissue specimens by matrix-assisted laser desorption/ionization imaging mass spectrometry in biological and clinical research. Chem. Rev. 113, 2309–2342 (2013)
Angel, P.M., Spraggins, J.M., Baldwin, H.S., Caprioli, R.: Enhanced sensitivity for high spatial resolution lipid analysis by negative ion mode matrix assisted laser desorption ionization imaging mass spectrometry. Anal. Chem. 84, 1557–1564 (2012)
Shariatgorji, M., Kallback, P., Gustavsson, L., Schintu, N., Svenningsson, P., Goodwin, R.J., Andren, P.E.: Controlled-pH tissue cleanup protocol for signal enhancement of small molecule drugs analyzed by MALDI-MS imaging. Anal. Chem. 84, 4603–4607 (2012)
Aerni, H.R., Cornett, D.S., Caprioli, R.M.: Automated acoustic matrix deposition for MALDI sample preparation. Anal. Chem. 78, 827–834 (2006)
Hankin, J.A., Barkley, R.M., Murphy, R.C.: Sublimation as a method of matrix application for mass spectrometric imaging. J. Am. Soc. Mass Spectrom. 18, 1646–1652 (2007)
Puolitaival, S.M., Burnum, K.E., Cornett, D.S., Caprioli, R.M.: Solvent-free matrix dry-coating for MALDI imaging of phospholipids. J. Am. Soc. Mass Spectrom. 19, 882–886 (2008)
Cornett, D.S., Reyzer, M.L., Chaurand, P., Caprioli, R.M.: MALDI imaging mass spectrometry: molecular snapshots of biochemical systems. Nat. Methods 4, 828–833 (2007)
Franck, J., Arafah, K., Barnes, A., Wisztorski, M., Salzet, M., Fournier, I.: Improving tissue preparation for matrix-assisted laser desorption ionization mass spectrometry imaging. Part 1: using microspotting. Anal. Chem. 81, 8193–8202 (2009)
Grove, K.J., Frappier, S.L., Caprioli, R.M.: Matrix precoated MALDI MS targets for small molecule imaging in tissues. J. Am. Soc. Mass Spectrom. 22, 192–195 (2011)
Yang, J., Caprioli, R.M.: Matrix precoated targets for direct lipid analysis and imaging of tissue. Anal. Chem. 85, 2907–2912 (2013)
Gemperline, E., Rawson, S., Li, L.: Optimization and comparison of multiple MALDI matrix application methods for small molecule mass spectrometric imaging. Anal. Chem. 86, 10030–10035 (2014)
Deutskens, F., Yang, J., Caprioli, R.M.: High spatial resolution imaging mass spectrometry and classical histology on a single tissue section. J. Mass Spectrom. 46, 568–571 (2011)
Castellino, S., Groseclose, M.R., Wagner, D.: MALDI imaging mass spectrometry: bridging biology and chemistry in drug development. Bioanalysis 3, 2427–2441 (2011)
Chaurand, P., Schwartz, S.A., Billheimer, D., Xu, B.J., Crecelius, A., Caprioli, R.M.: Integrating histology and imaging mass spectrometry. Anal. Chem. 76, 1145–1155 (2004)
Hsieh, Y., Casale, R., Fukuda, E., Chen, J., Knemeyer, I., Wingate, J., Morrison, R., Korfmacher, W. : Matrix-assisted laser desorption/ionization imaging mass spectrometry for direct measurement of clozapine in rat brain tissue. Rapid Commun. Mass Spectrom. 20, 965–972 (2006)
Acknowledgments
The authors acknowledge the thoughtful discussions and support received from Joey C. Latham, Edna Choo, and Justin Ly as well as, from members of the Drug Metabolism and Pharmacokinetics, IVS and Formulation Departments at Genentech, Inc. Additionally, authors express their gratitude to Katherine Kellersberger, Christian Berg, and Bill Engar of Bruker Daltonics.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplemental Figure 1
(PPTX 623 kb)
Rights and permissions
About this article
Cite this article
Quiason, C.M., Shahidi-Latham, S.K. Imaging MALDI MS of Dosed Brain Tissues Utilizing an Alternative Analyte Pre-extraction Approach. J. Am. Soc. Mass Spectrom. 26, 967–973 (2015). https://doi.org/10.1007/s13361-015-1132-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13361-015-1132-z