Abstract
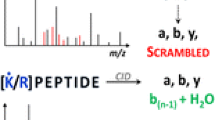
An understanding of the process of peptide fragmentation and what parameters are best to obtain the most useful information is important. This is especially true for large-scale proteomics where data collection and data analysis are most often automated, and manual interpretation of spectra is rare because of the vast amounts of data generated. We show herein that collisional cell peptide fragmentation, in this case higher collisional dissociation (HCD) in the Q Exactive, is significantly affected by the normalized energy applied. Both peptide sequence and energy applied determine what ion fragments are observed. However, by applying a stepped normalized collisional energy scheme and combining ions from low, medium, and high collision energies, we are able to increase the diversity of fragmentation ions generated. Application of stepped collision energy to HEK293T lysate demonstrated a minimal effect on peptide and protein identification in a large-scale proteomics dataset, but improved phospho site localization through increased sequence coverage. Stepped HCD is also beneficial for tandem mass tagged (TMT) experiments, increasing intensity of TMT reporters used for quantitation without adversely effecting peptide identification.

ᅟ






Similar content being viewed by others
References
Frank, A.M.: Predicting intensity ranks of peptide fragment ions. J. Proteome Res. 8(5), 2226–2240 (2009)
Yen, C.-Y., Meyer-Arendt, K., Eichelberger, B., Sun, S., Houel, S., Old, W.M., Knight, R., Ahn, N.G., Hunter, L.E., Resing, K.A.: A Simulated MS/MS library for spectrum-to-spectrum searching in large scale identification of proteins. Mol. Cell. Proteomics. 8(4), 857–869 (2009)
Biemann, K.: Sequencing of peptides by tandem mass-spectrometry and high-energy collision-induced dissociation. Methods Enzymol. 193, 455–479 (1990)
Hunt, D.F., Yates, J.R., Shabanowitz, J., Winston, S., Hauer, C.R.: Protein sequencing by tandem mass-spectrometry. Proc. Natl. Acad. Sci. U. S. A. 83(17), 6233–6237 (1986)
de Graaf, E.L., Altelaar, A.F.M., van Breukelen, B., Mohammed, S., Heck, A.J.R.: Improving SRM assay development: a global comparison between triple quadrupole, ion trap, and higher energy CID peptide fragmentation spectra. J. Proteome Res. 10(9), 4334–4341 (2011)
Michalski, A., Damoc, E., Hauschild, J.-P., Lange, O., Wieghaus, A., Makarov, A., Nagaraj, N., Cox, J., Mann, M., Horning, S.: Mass spectrometry-based proteomics using Q Exactive, a high-performance benchtop quadrupole Orbitrap mass spectrometer. Mol. Cell. Proteom. 10(9), M111.011015 (2011)
Kelstrup, C.D., Young, C., Lavallee, R., Nielsen, M.L., Olsen, J.V.: Optimized fast and sensitive acquisition methods for shotgun proteomics on a quadrupole Orbitrap mass spectrometer. J. Proteome Res. 11(6), 3487–3497 (2012)
Mayer, P.M., Poon, C.: The mechanisms of collisional activation of ions in mass spectrometry. Mass Spectrom. Rev. 28(4), 608–639 (2009)
Michalski, A., Neuhauser, N., Cox, J., Mann, M.: A systematic investigation into the nature of tryptic HCD spectra. J. Proteome Res. 11(11), 5479–5491 (2012)
van Dongen, W.D., Ruijters, H.F.M., Luinge, H.J., Heerma, W., Haverkamp, J.: Statistical analysis of mass spectral data obtained from singly protonated peptides under high-energy collision-induced dissociation conditions. J. Mass Spectromics. 31(10), 1156–1162 (1996)
Cox, K.A., Gaskell, S.J., Morris, M., Whiting, A.: Role of the site of protonation in the low-energy decompositions of gas-phase peptide ions. J. Am. Soc. Mass Spectrom. 7(6), 522–531 (1996)
Dongre, A.R., Jones, J.L., Somogyi, A., Wysocki, V.H.: Influence of peptide composition, gas-phase basicity, and chemical modification on fragmentation efficiency: evidence for the mobile proton model. J. Am. Chem. Soc. 118(35), 8365–8374 (1996)
Jedrychowski, M.P., Huttlin, E.L., Haas, W., Sowa, M.E., Rad, R., Gygi, S.P.: Evaluation of HCD- and CID-type Fragmentation within their respective detection platforms for murine phosphoproteomics. Mol. Cell Proteomics 10(12), M111.009910 (2011)
Frese, C.K., Altelaar, A.F.M., Hennrich, M.L., Nolting, D., Zeller, M., Griep-Raming, J., Heck, A.J.R., Mohammed, S.: Improved peptide identification by targeted fragmentation using CID, HCD, and ETD on an LTQ-Orbitrap Velos. J. Proteome Res. 10(5), 2377–2388 (2011)
Shen, Y.F., Tolic, N., Purvine, S.O., Smith, R.D.: Improving collision induced dissociation (CID), high energy collision dissociation (HCD), and electron transfer dissociation (ETD) Fourier tTransform MS/MS degradome-peptidome identifications using high accuracy mass information. J. Proteome Res. 11(2), 668–677 (2012)
Nagaraj, N., D'Souza, R.C.J., Cox, J., Olsen, J.V., Mann, M.: Feasibility of Large-scale phosphoproteomics with higher energy collisional dissociation fragmentation. J. Proteome Res. 9(12), 6786–6794 (2010)
Bushee, J.L., Argikar, U.A.: An experimental approach to enhance precursor ion fragmentation for metabolite identification studies: application of dual collision cells in an orbital trap. Rapid Commun. Mass Spectrom. 25(10), 1356–1362 ( 2011)
Chakraborty, A.B., Berger, S.J., Gebler, J.C.: Use of an integrated MS-multiplexed MS/MS data acquisition strategy for high-coverage peptide mapping studies. Rapid Commun. Mass Spectrom. 21(5), 730–744 (2007)
Dayon, L., Pasquarello, C., Hoogland, C., Sanchez, J.C., Scherl, A.: Combining low- and high-energy tandem mass spectra for optimized peptide quantification with isobaric tags. J. Proteom. 73(4), 769–777 (2010)
McDonald, W.H., Tabb, D.L., Sadygov, R.G., MacCoss, M.J., Venable, J., Graumann, J., Johnson, J.R., Cociorva, D., Yates, J.R.: MS1, MS2, and SQT—three unified, compact, and easily parsed file formats for the storage of shotgun proteomic spectra and identifications. Rapid Commun. Mass Spectrom. 18(18), 2162–2168 (2004)
Tabb, D.L., McDonald, W.H., Yates, J.R.: DTASelect and contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J. Proteome Res. 1(1), 21–26 (2002)
Falick, A.M., Hines, W.M., Medzihradszky, K.F., Baldwin, M.A., Gibson, B.W.: Low-mass ions produced from peptides by high-energy collision-induced dissociation in tandem mass-spectrometry. J. Am. Soc. Mass Spectrom. 4(11), 882–893 (1993)
Tabb, D.L., MacCoss, M.J., Wu, C.C., Anderson, S.D., Yates, J.R.: Similarity among tandem mass spectra from proteomic experiments: detection, significance, and utility. Anal. Chem. 75(10), 2470–2477 (2003)
Jue, A.L., Racine, A.H., Glish, G.L.: The effect of ion trap temperature on the dissociation of peptide ions in a quadrupole ion trap. Int. J. Mass Spectrom. 301(1–3), 74–83 (2011)
Yalcin, T., Csizmadia, I.G., Peterson, M.R., Harrison, A.G.: The structure and fragmentation of B-n (n ≥ 3) ions in peptide spectra. J. Am. Soc. Mass Spectrom. 7(3), 233–242 (1996)
Papayannopoulos, I. A.: The interpretation of collision-induced dissociation tandem mass-spectra of peptides. Mass Spectrom. Rev. 14(1), 49–73 (1995)
Eng, J.K., McCormack, A.L., Yates, J.R.: An approach to correlate tandem mass-spectral data of peptides with amino-acid-sequences in a protein database. J. Am. Soc. Mass Spectrom. 5(11), 976–989 (1994)
Paizs, B., Szlavik, Z., Lendvay, G., Vekey, K., Suhai, S.: Formation of a2+ ions of protonated peptides. An ab initio study. Rapid Commun. Mass Spectrom. 14(9), 746–755 (2000)
Olsen, J.V., Blagoev, B., Gnad, F., Macek, B., Kumar, C., Mortensen, P., Mann, M.: Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127(3), 635–648 (2006)
Steen, H., Kuster, B., Fernandez, M., Pandey, A., Mann, M.: Detection of tyrosine phosphorylated peptides by precursor ion scanning quadrupole TOF mass spectrometry in positive ion mode. Anal. Chem. 73(7), 1440–1448 (2001)
Olsen, J.V., Macek, B., Lange, O., Makarov, A., Horning, S., Mann, M.: Higher-energy C-trap dissociation for peptide modification analysis. Nat. Methods 4(9), 709–712 (2007)
Johnson, R.S., Martin, S.A., Biemann, K., Stults, J.T., Watson, J.T.: Novel fragmentation process of peptides by collision-induced decomposition in a tandem mass-spectrometer—differentiation of leucine and isoleucine. Anal. Chem. 59(21), 2621–2625 (1987)
Johnson, R.S., Martin, S.A., Biemann, K.: Collision-induced fragmentation of (M+H)+ ions of peptides. Side chain specific sequence ions. Int. J. Mass Spectrom. Ion Process. 86, 137–154 (1988)
Acknowledgments
The authors thank Aaron Aslanian for culture of HEK cells, and Claire Delahunty for the critical reading of the manuscript. Financial support was provided by NIH grants P41 RR011823/GM103533, R01 MH067880-09, P01 AG031097-03, and R01 HL079442-07.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Diedrich, J.K., Pinto, A.F.M. & Yates, J.R. Energy Dependence of HCD on Peptide Fragmentation: Stepped Collisional Energy Finds the Sweet Spot. J. Am. Soc. Mass Spectrom. 24, 1690–1699 (2013). https://doi.org/10.1007/s13361-013-0709-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13361-013-0709-7