Abstract
Benzoxazinoids (BX) are major secondary metabolites of gramineous plants that play an important role in disease resistance and allelopathy. They also have many other unique properties including anti-bacterial and anti-fungal activity, and the ability to reduce alfa–amylase activity. The biosynthesis and modification of BX are controlled by the genes Bx1 ÷ Bx10, GT and glu, and the majority of these Bx genes have been mapped in maize, wheat and rye. However, the genetic basis of BX biosynthesis remains largely uncharacterized apart from some data from maize and wheat. The aim of this study was to isolate, sequence and characterize five genes (ScBx1, ScBx2, ScBx3, ScBx4 and ScBx5) encoding enzymes involved in the synthesis of DIBOA, an important defense compound of rye. Using a modified 3D procedure of BAC library screening, seven BAC clones containing all of the ScBx genes were isolated and sequenced. Bioinformatic analyses of the resulting contigs were used to examine the structure and other features of these genes, including their promoters, introns and 3’UTRs. Comparative analysis showed that the ScBx genes are similar to those of other Poaceae species, especially to the TaBx genes. The polymorphisms present both in the coding sequences and non-coding regions of ScBx in relation to other Bx genes are predicted to have an impact on the expression, structure and properties of the encoded proteins.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Benzoxazinoids (BX) are protective and allelopathic secondary metabolites found in a large number of species belonging to the Poaceae family, including the major agricultural cereals maize, wheat and rye (Frey et al. 2009; Niemeyer 2009). The properties and biosynthesis of BX have been intensively studied for over 50 years; they were first discovered and characterized in rye (Virtanen and Hietala 1955a, b), wheat and maize (Wahlroos and Virtanen 1959) in the 1950s. The first step in BX biosynthesis in maize, diploid and hexaploid wheat, and Hordeum lechleri, is the conversion of indole-3-glycerolphosphate to indole that occurs in chloroplasts. The products of the subsequent four reactions, taking place in endoplasmatic reticulum, are the four main BXs: HBOA (2-hydroxy-1,4-benzoxazin-3-one), DIBOA (2,4-dihydroxy-1,4-benzoxazin-3-one), TRIBOA (2,4,7-trihydroxy-1,4-benzoxazin-3-one) and DIMBOA (2,4-dihydroxy-7-methoxy-1,4-benzoxazin-3-one). The final products of BX biosynthesis are the glycosides, which are stored in the vacuole (Frey et al. 2009; Niemeyer 2009). Upon disintegration of the cell due to pathogen or pest attack and the mobilization of jasmonic acid and/or its methyl ester, glycosidases stored in chloroplasts are activated and toxic BX aglucons are produced (Oikawa et al. 2002; Niemeyer 2009).
DIMBOA is the main aglucon in maize and wheat, whereas in rye DIBOA and its more stable degradation product BOA (2-benzoxazolin-2(3H)-one) are predominant. DIBOA was found to be the major compound in rye leaves, while both DIMBOA and DIBOA were identified in rye roots (Frey et al. 2009; Niemeyer 2009). DIBOA-Glc is the final product of BX biosynthesis in wild Hordeum species and several other species of Poaceae (Frey et al. 2009). This compound has also been detected in leaf and root extracts of rye (Zasada et al. 2007; Meyer et al. 2009).
Several genes controlling BX biosynthesis have been isolated and characterized. The enzymes participating in BX biosynthesis in maize are encoded by the genes ZmBx1 ÷ ZmBx10: Bx1 – indole-3-glycerol phosphate lyase; Bx2 ÷ Bx5 – cytochrome P450 monooxygenases, members of the CYP71 family; Bx6 – 2-oxoglutarate dependent dioxygenase; Bx7 – 7-O-methyltransferase; Bx8, Bx9 – UDP-glucosyltransferases; Bx10a,b,c – 4-O-methyltransferases (Jonczyk et al. 2008; Frey et al. 2009; Meihls et al. 2013). The genes Bx1 to Bx5 have also been isolated from hexaploid (Triticum aestivum) and diploid wheat (Triticum monococcum, Triticum urartu and Triticum boeoticum), (Nomura et al. 2003; Jonczyk et al. 2008; Niemeyer 2009; Frey et al. 2009). The Bx6 and Bx7, encoding enzymes that catalyze the sequential 7-hydroxylation and 7-O-methylation of DIBOA-Glc to DIMBOA-Glc, have been identified in maize, but not in wheat or rye, although the same reactions probably occur in these cereals (Frey et al. 2009; Sue et al. 2011). Indeed, the mRNA of a Bx6-like gene coding for 2,4-dihydroxy-1,4-benzoxazin-3-one-glucoside dioxygenase has recently been described in rye (http://www.ncbi.nlm.nih.gov/nuccore/HG380515.1÷520.1). The genes ZmBx1 ÷ ZmBx8 are clustered and located on the short arm of maize chromosome 4, ZmBx9/GT, which are functionally almost identical to ZmBx8 and ZmBx10a,b,c are on chromosome 1, whereas Zmglu1 and Zmglu2, encoding glucosideglucosidases, are on chromosome 10. In hexaploid wheat, the Bx gene cluster is divided between groups: 2 – glu homologs, 4 – TaBx1 and TaBx2, 5 – TaBx3 ÷ TaBx5, and 7 – GT homologs (Jonczyk et al. 2008; Frey et al. 2009; Niemeyer 2009; Sue et al. 2011). The majority of Bx genes identified so far have been sequenced, at least at the cDNA level.
In rye, homeoloci of TaBx1 and TaBx2 were identified on chromosome 7R (ScBx1 and ScBx2), and those of TaBx3 ÷ TaBx5 on chromosome 5R (ScBx3 ÷ ScBx5), (Nomura et al. 2003, 2008; Frey et al. 2009; Niemeyer 2009). Recently, Sue et al. (2011) showed that two rye Bx genes, ScGT, an ortholog of ZmBx8/ZmBx9, and Scglu, an ortholog of Zmglu1 and Zmglu2, are located on chromosomes 4R and 2R, respectively. The cDNA sequences of six Bx genes of rye are available: ScBx1 ÷ ScBx5 and a Bx6-like gene (La Hovary 2012, http://www.ncbi.nlm.nih.gov/nuccore/HG380515.1 ÷520.1).
The aims of this study were to examine the sequences of the full-length rye ScBx1 ÷ ScBx5 genes in order to characterize their exons, introns, UTRs and promoters, to compare their structures with Bx genes from other species, and to predict their likely role based on promoter analysis.
Materials and methods
Plant material and DNA isolation
DNA was isolated from young seedlings of winter rye (Secale cereale L.) inbred line L318 (S20–S22) using the CTAB method (Murray and Thompson 1980). BAC clone DNA was isolated using a modified alkaline lysis method and pooled using the three-dimensional (3D) procedure recommended by Amplicon Express, described below (Isolation of positive BAC clones section). The DNA concentration was measured using a NanoDrop 2000 spectrophotometer.
Primer design and PCR
Specific primers for genes ScBx1 and ScBx2 were designed based on the rye cDNA and mRNA sequences (GenBank accession no. JQ716987.1 and JX442061.1, respectively), while for the other ScBx genes, the sequences of mRNAs of Triticum aestivum (B genome) were used. Rye line L318-specific primers were designed based on two selected amplicons per gene. In total, ten primer pairs were used for BAC library screening (Table 1).
PCRs were composed of 500 ng total genomic DNA, 3 μM F and R primers, 0.2 mM dNTPs, 0.5 mM MgCl2, 1x PCR buffer and 3 units of DreamTaq polymerase (Fermentas) in a total volume of 15 μl. Amplification was performed in a thermal cycler using the following conditions: (1) 94 °C for 1 min; (2) 94 °C for 30 s, 60 °C for 30 s, 72 °C for 60 s for 35 cycles; (3) 72 °C for 5 min. The products were separated on a 1 % agarose gel, stained with ethidium bromide and visualized on a UV transilluminator.
Cloning, sequencing and BLAST analysis
All amplicons were purified using a GeneJET PCR Purification Kit (Thermo Scientific) and sequenced by a commercial sequencing company (Genomed S.A., Warsaw). The resulting sequences were compared with those of all Bx orthologs available in databases using the BLAST algorithm.
Construction of a BAC library
For the preparation of high molecular weight (HMW) rye DNA, nuclei were isolated and purified by flow cytometry as described by Šimková et al. (2003). Approximately 48,000 nuclei (corresponding to ca. 0.8 μg DNA) were embedded in 80 μl agarose plugs and DNA isolation was performed according to Šimková et al. (2003), except that the plugs were washed six times in ice-cold TE buffer before digestion. A BAC library was constructed as described by Peterson et al. (2000) with some modifications. Each plug was cut into nine pieces, which were divided among three tubes. For partial digestion of the HMW DNA, 0.4 to 1.2 U of HindIII (0.7 U on average) were used per tube. Two rounds of size selection were then performed. In the first round, the partially digested DNA was resolved on a 1 % SeaKem Gold Agarose gel (Lonza, Rockland, USA) in 0.25xTBE by pulsed-field gel electrophoresis (PFGE) using the following conditions: voltage 6 V/cm, switch time 1–50 s, run time 17 h. The size fraction of 100–150 kb was cut from the gel and subjected to a second round of electrophoretic size selection on a 0.9 % SeaKem Gold Agarose gel in 0.25xTBE using the following conditions: voltage 6 V/cm, switch time 3 s, run time 17 h. The 100–150-kb size fraction was cut from the gel and subdivided into two fractions of 100–120 kb (B) and 120–150 kb (M). The DNA in each fraction was electroeluted from the gel and quantified by electrophoresis on a standard 1 % agarose gel alongside a phage lambda DNA dilution series. The DNA from both of the fractions was ligated with HindIII-digested cloning-ready pIndigoBAC-5 vector (Epicentre, Madison, USA) using DNA:vector mass ratios of 3.75:1 for B and 4:1 for M. Escherichia coli ElectroMAX DH10B competent cells (Invitrogen, Carlsbad, USA) were then transformed with the ligations. The resulting BAC clone library, comprised of 105,216 individual clones, was ordered in 384-well plates filled with 75 μl of freezing medium (2YT supplemented with 6.6 % glycerol and 12.5 mg/l chloramphenicol) using a Qbot (Genetix, New Milton, UK) and stored at −80 °C.
Isolation of positive BAC clones
The isolation of BAC clones containing ScBx genes was based on the Amplicon Express strategy (http://ampliconexpress.com/products-services/screening-services/pools-and-superpools).
Briefly, 39 superpools, each containing 2688 individual BAC clones from seven plates were prepared for the first round of PCR. The second PCR round was performed on the matrixed Plate, Row and Column pools from the selected Superpools. Finally, the “positive” BAC clones containing the desired sequences were picked and sequenced (Genomed S.A.).
Bioinformatic analyses
Bioinformatic analyses were performed by means of the programs listed below using default settings except for SoftBerry/FGENESH when monocot plant specific gene-finding parameters and Blastn when the selection of nucleotide collection (in “other databases” section) and megablast (highly similar sequences) algorithm were applied:
-
For ScBx genes/amplicons preliminary identification: Blastn (www.ncbi.nlm.nih.gov/)
-
For constructing contigs from short amplicons after obtained in PCR during the preliminary studies: Sequencher 4.5
-
For identifing Bx genes in the total sequence of all BAC contigs: BioEdit ver. 7.0.9.0
-
For ScBx genes structure (exon, intron and end of 3’UTR positions within ScBx genes) determination: SoftBerry/FGENESH (http://www.softberry.com). Additionally, ClustalW2 (http://www.ebi.ac.uk/Tools/msa/clustalw2/) and Emboss tools: needle, seqcut, showseq (http://emboss.bioinformatics.nl/) were used as supporting tools to gene assembling
-
For promoter analysis: PlantCare (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/, Lescot et al. 2002). The possibly longest sequences (except for ScBx1, where a length of 3000 bp was arbitrarily selected) of predicted ScBx promoters were scanned for the presence of putative cis-acting regulatory elements. For stress-specific motifs (SSM) identifying, the “search for care” query was used. The frequency of SSM was calculated according to the formula: total number of given types of SSM divided by the total analyzed sequence upstream the start codon x 100
-
For protein structure and function prediction: I-TASSER (http://zhanglab.ccmb.med.umich.edu/I-TASSER/, Zhang 2008; Roy et al. 2010). The cDNA sequences were changed into amino acid sequences and put into I-TASSER analysis. Then from five models, the highest scoring I-TASSER model, based on C-score,Footnote 1 was chosen
Phylogenetic trees were generated separately for the cDNA sequences of the Bx1 and Bx2 ÷ Bx5 genes using Mega6 software (Tamura et al. 2013) based on the Maximum Parsimony algorithm (Nei and Kumar 2000) with bootstrap consensus from 2000 replicates (Felsenstein 1985). The alignments used to build phylogenetic trees were done in Mega6 software. In the first step, cDNA sequences from GenBank were translated into aminoacid sequences followed by alignment done by means of ClustalW (attached to Mega6). After that, the aminoacid sequences were transformed again into cDNA sequences and used for further phylogenetic analysis. In the case of Bx1 gene, one out of two generated trees was chosen; for Bx2 ÷ Bx5 genes only one tree was computed.
In the text, the ScBx genes isolated in this study are named KF- ScBx, while the genes isolated by La Hovary (2012) are referred to as J- ScBx and those isolated by Tanwir et al. (http://www.ncbi.nlm.nih.gov/nuccore/HG380515.1 ÷520.1) from cv. Picasso, as HG- ScBx.
Results and discussion
Although the biosynthesis of benzoxazinoids has been intensively studied for over 50 years, little is known about the genes involved and the mechanisms controlling their differential expression in different plant tissues and during plant ontogeny. Maize, wheat and Hordeum lechleri are the best characterized species with regard to the genetic basis of BX synthesis. Prior to this study, the only available genetic data relating to BX synthesis in rye were the genomic location of ScBx1 ÷ ScBx5 gene orthologs and the coding sequences of ScBx1 and ScBx2 genes. The construction of a BAC library (Šimková H and Rakoczy-Trojanowska 2010) enabled the isolation and precise characterization of these five genes controlling BX biosynthesis, including their introns and promoters, and permitted phylogenetic analysis.
Isolation of positive BAC clones
Using PCR with rye KF-ScBx1 ÷ ScBx5 gene-specific primers, seven clones were isolated from the rye inbred line L318 (S20) BAC library (105,216 clones). These clones were sequenced and the resulting reads were aligned in contigs. Altogether, 93 contigs were arranged: between 6 and 25 per BAC clone (Table 2). The desired ScBx sequences were identified in all selected clones. Except for clones 2 and 4, where only single ScBx sequences were found (ScBx5 and ScBx3, respectively), two or three ScBx genes were present in the remaining clones. Some of the gene pairs ScBx1-ScBx2 and ScBx3-ScBx4 were present in the same contigs, whereas ScBx5 was found in separate ones. The isolation efficiency was 0.0007 % and the presence of a given ScBx gene in more than one BAC clone indicated that ScBxs are present in the rye genome as single-copy genes. Apart from the detection of an additional TaBx3 copy on the long arm of chromosome 5B (Nomura et al. 2003), there are no data indicating the existence of more than one copy of any Bx1, Bx2, Bx4 and Bx5 gene per genome.
Characteristics of the KF-ScBx1 ÷ ScBx5 genes
Structure
The predicted structure and length of the five KF-ScBx genes are presented in Table 3 and Fig. 1. The gene ScBx1 is composed of seven exons and six introns, ScBx2 has two exons and one intron, and each of the genes ScBx3 ÷ ScBx5 has three exons and two introns. While ScBx1 has the largest number of exons, the length of individual exons and their total length are the lowest among all the ScBx genes. Despite the fact that ScBx2 contains two exons and ScBx3 ÷ ScBx5 have three exons each, the total exon length of these ScBx genes is very similar. The ScBx5 gene has the longest sequence due to the possession of relatively long introns (more than twice longer than in the genes ScBx2 ÷ ScBx4) and an unusually long 3′UTR. It was not possible to characterize the 5′UTR of the newly isolated rye Bx genes by bioinformatic analysis alone. The position and number of introns differ between genes KF-ScBx2 ÷ ScBx5: ScBx2 has only one, most probably the ancestral intron, whereas ScBx3, ScBx4 and ScBx5 – two introns (the ancestral intron 2 and the intron 1 which was gained after the first duplication in the CYP71C subfamily as hypothesized by Dutartre et al. 2012). However, their positions differ from that suggested by Dutartre et al. (2012) which calls in question the hypothesis about conserved positions of introns within Bx2 ÷ Bx5 clades in the whole Poaceae family (for more details see supplementary materials, Table 4). The observation that the introns 1 and 2 are positioned identically or similarly confirms the monophyletic origin of ScBx3 ÷ ScBx5 genes.
Predicted function
Use of the ScBx gene sequences to search the NCBI database (http://blast.ncbi.nlm.nih.gov/Blast.cgi) identified highly significant similarities between the coding sequences (formed by splicing the exons together) of the 5 KF-ScBx genes and the Bx genes of rye (HG and J sequences), H. lechleri, Z. mays and three genomes of T. aestivum (for more details see supplementary materials, Table 1). All of the KF genes, apart from ScBx1, are most similar to the rye and wheat genes, and the least similar to the maize genes. In contrast, the KF-ScBx1 sequence is more similar to the equivalent Z. mays gene than to that of H. lechleri. A more detailed comparison is described in Comparative structural analysis of Bx genes subsection.
Based on the results of our bioinformatic analysis, the proteins encoded by the KF-ScBx genes probably have the following functions:
-
ScBx1 – indole-3-glycerol phosphate lyase
-
ScBx2 – indole monooxygenase
-
ScBx3 – indolin-2-one monooxygenase
-
ScBx4 – 3-hydroxyindolin-2-one monooxygenase
-
ScBx5 – 2-hydroxy-1,4-benzoxazin-3-one monooxygenase
Arrangement
It was possible to establish the arrangement of gene pairs ScBx1 ÷ ScBx2 and ScBx3 ÷ ScBx4 since they were present in the same contig(s) of given BAC clones: ScBx1 ÷ ScBx2 in contig 1 of clone 5, and ScBx3 ÷ ScBx4 in contig 6 of clone 3 and contig 4 of clone 6 (Table 2). The distances (from the last nucleotide of the 3′UTR of the first gene to the first nucleotide of the first exon of the second gene in a given pair) between genes ScBx1 and ScBx2, organized “tail-to-head”, is 1960 bp and between ScBx3 and ScBx4, also arranged “tail-to-head”, is 8073 bp. This arrangement of these two pairs of genes is like that found in Triticum aestivum (Nomura et al. 2003, 2005; Frey et al. 1997; Jonczyk et al. 2008). The distance between ScBx3 and ScBx4 is also similar to that of the wheat orthologs (8974, 7255 and 11,309 bp in the A, B and D genomes, respectively), but considerably less than that in maize (45,767 bp). In the case of the gene pair ScBx1 ÷ ScBx2, it is only possible to compare the separation distance with that in maize: ZmBx1 is 2490 bp downstream of ZmBx2 (Frey et al. 1997). We speculate that this spacing is similar in wheat because both deletion mapping and Southern hybridization analysis revealed that TaBx1 and TaBx2 are located close together in the same region of each chromosome (Nomura et al. 2003), but no supporting experimental evidence is available.
Since ScBx5 was not found in the same contig as ScBx3 and ScBx4, it was not possible to determine the distances between these three genes. Because these genes were always present in the same BACs, it may be concluded that, as in wheat (Nomura et al. 2003, 2005; Sue et al. 2011), the KF-ScBx3 ÷ ScBx5 genes are located on the same chromosome (5 RS). Although we cannot precisely determine the arm locations of KF-ScBx3 ÷ ScBx5, or their separation distances in the rye genome, an approximate evaluation is possible based on the ordering and lengths of contigs from BACs 3 and 6. The spaces between ScBx5 and ScBx3, and between ScBx5 and ScBx4 are at least 2929 bp and 14,353 bp, respectively.
Promoters
In all of the analyzed promoters, SSM potentially involved in different abiotic (e.g., heat or drought) and biotic (e.g., fungal elicitor) stress response reactions, as well as in signaling pathway mobilization (e.g., MeJa- or SA-mediated pathways), were found (Table 4). The sequences of cis-acting regulatory elements involved in MeJA-responsiveness (CGTCA and TGACG) were those most frequently detected. The highest frequency of SSMs was in the promoters of genes ScBx3 and ScBx4: 0.74 and 1.15 SSM per 100 nt, respectively. The promoter binding sites MYB and MYBHv1 (analogous to human myeloblastosis gene family coding for transcription factor Myb), typical for the majority of stress-related gene promoters, were identified in almost all of the analyzed promoters except that of ScBx4.
A comparison of the KF-ScBx promoters with those of the genes TaBx3, TaBx4 and TaBx5 (sequences of ≥ 200 nt are only available for these three genes and ZmBx1 ÷ ZmBx5, showed that the majority of SSMs are common to all three species; for more details see supplementary materials, Table 2). However, some potentially important differences were found. The cis-acting element involved in abscisic acid responsiveness, present in some wheat (TaBx3, TaBx4) and maize (ZmBx1, ZmBx3, ZmBx5) promoters, was not identified in rye. Other SSM regulatory elements involved in cold- and dehydration-responsiveness were only detected in wheat. A cis-acting regulatory element involved in MeJA-responsiveness was present in rye and maize, but not in wheat. These findings suggest that the analyzed rye Bx genes play roles in different stress response reactions and are consistent with the existing knowledge. Benzoxazinoids, except for their allelopathic potential are proved to be a crucial element in the defense mechanisms of many species belonging to the Poaceae family against biotic stresses as pests or pathogens (Niemeyer 2009). Their role in the defense against abiotic stresses (drought, soil salinity, triazine derivatives, aluminum) has also been documented (Makleit 2005; Kato-Noguchi 2008; Niemeyer 2009) but much more poorly and fragmentarily. In this context, an identification in ScBx gene promoters of the abiotic stress specific motifs is of spectacular importance. Nevertheless, the specificity of some rye Bx genes, especially ScBx3 and ScBx4, appears to be slightly different to that of the equivalent genes of wheat and maize or rye SSMs differ from wheat/maize ones. Moreover, until the experimental evidence is available, we can only predict, on the base of promoter bioinformatic analysis, the function of ScBx genes.
The frequency of SSM elements in the compared promoters of the Bx3 and Bx4 genes was much lower in wheat and maize than in rye amounting: 0.27, 0.67, 0.24 and 0.31 for TaBx3, TaBx4, ZmBx3 and ZmBx4, respectively, and it was comparable in the case of Bx5 genes (for more details see supplementary materials, Table 2a, 2b). For contrast, we have also compared the frequency of potential SSMs between ScBx gene promoters and the total contig sequence (806,758 bp) of seven BAC clones which included ScBx genes (Table 2) which turned to be considerably lower, at least fourfold, than in promoters. Some motifs — occurred either sporadically — AAAAAATTTC (for heat stress responsiveness), CCATCTTTTT and CAGAAAAGGA (for salicylic acid responsiveness), ATTTTCTTCA and ATTTTCTCCA (for defense and stress responsiveness), and some motifs: AGAAAATTCG (for heat stress responsiveness) and GTTTTCTTAC (for defense and stress responsiveness) were not present at all (for more details see supplementary materials, Table 2c).
Comparative structural analysis of Bx genes
ScBx genes of different rye accessions
The availability of only incomplete data prevented full comparison of the KF-ScBx genes with those from accessions HG and J. The coding sequences of the genes ScBx1 and ScBx3 were the same in different rye accessions, but slight differences were found in the coding sequences of the other genes and in the 3′UTR lengths (Table 5).
All ScBx genes from the different rye accessions were similar at the nucleotide level (for more details see supplementary materials, Table 3), but only the ScBx3 gene sequences were identical. The other ScBx genes varied by the presence of a few SNPs and INDELs. The highest number of SNPs was identified in the first exon of ScBx2, with A/G, C/T and T/C being the most frequent polymorphisms. Of the 19 identified SNPs, three caused non-conserved and one a semi-conserved substitution. Three INDELs caused length mismatches between the ScBx2, ScBx4 and ScBx5 genes of the KF and HG accessions. These are present in the first exon of KF-ScBx2 (24-bp deletion: ATGGCTCAGGTACATGTAGAAGAG), the first exon of KF-ScBx4 (57-bp insertion: GCAGCGCGCCGTCGGCCATGGCGTATCCACAGAAGCACTACTGCT…CT) and the second exon of KF-ScBx5 (3-bp deletion: TCC), (for more details see supplementary materials, Table 3).
The non-conserved amino acid (AA) substitutions and INDELs may have an impact on protein properties. To test this hypothesis, we performed bioinformatic prediction of the changes in protein structure caused by two major INDELs present in two of the KF-ScBx genes: a 24-bp deletion in the first exon of KF-ScBx2 and a 57-bp insertion in the first exon of KF-ScBx4 (for more details see supplementary materials, Fig. 1 a, b). This analysis showed that these INDELs caused changes in the length of the first predicted α-helices of KF-ScBx2 (reduced) and KF- ScBx4 (increased) compared with those of the corresponding proteins of accession HG. No other structural alterations were found.
Bx genes in different cereal species
Maize, wheat and Hordeum lechleri are the best characterized species with regard to the genetic basis of BX synthesis (Frey et al. 1995, 1997, 2009; Jonczyk et al. 2008; Nomura et al. 2003, 2005; Grün et al. 2005). Therefore, we performed a comparative analysis including Bx genes from these species. However, in the case of wheat Bx1, Bx2 and Bx5, and H.lechleri Bx1 ÷ Bx5 the number of exons and introns were established bioinformatically as only cds sequences of these genes are available.
The majority of sequenced Bx genes had the same number of exons and introns in phase 0 as their orthologs. Two exceptions are the ZmBx1 gene with 6 exons, while this gene in all the other species has seven exons, and the ZmBx4 gene with only two exons and one, most probably ancestral, intron rather than three exons and two introns found in the equivalent Sc-, Ta- and HlBx genes (Table 6). The fifth and sixth exons of the rye and wheat Bx1 genes correspond to the fifth exon of the maize gene. The ZmBx4 intron matches the second intron of ScBx4, TaBx4 and HlBx4 genes.
These findings suggest that both the sixth exon of Sc/TaBx1 and the second intron of Sc/TaBx4 arose after differentiation into Panicoideae and Pooideae, and are consistent with the notion of a common, monophyletic origin of the BX biosynthetic pathway (e.g., Frey et al. 1997; Dutartre et al. 2012). However, the positions of introns in matched Bx2 ÷ Bx5 clades turned out to be highly conserved only for genes Bx3 and Bx4 of rye and wheat. In the case of the rest of genes we have noticed smaller or bigger differences; the most considerable dissimilarities were observed in comparisons with maize (for more details see supplementary materials, Table 4). Our results do not gainsay the hypothesis about the monophyletic origin of monooxygenase encoding genes but do not fully agree with the conception that the position of introns in the CYP71C family genes indicates their fully shared evolutionary origin as suggested by Dutartre et al. (2012).
Overall, the exon lengths of the KF-ScBx genes were very similar to those of the equivalent Bx genes of wheat and H. lechleri, but the exons of all maize Bx genes are slightly longer (Table 7). The lengths of the coding sequences of the genes Bx3 and Bx4 are identical in rye, wheat and H. lechleri, and are equal to 1584 bp and 1587 bp, respectively. The greatest variation in the Bx genes of different species is seen in the 3′UTR sequences. In the case of the gene ScBx5, the length of its predicted 3′UTR exceeds those of the other ScBx genes by more than four times.
As well as a general structural comparison of the KF-ScBx genes with TaBx, ZmBx and HlBx genes, we performed a more detailed analysis of polymorphisms present in both the coding and regulatory (when possible) components of these genes and the probable impact of the former on the structure and properties of the encoded proteins. A complete comparison was not possible because some of the necessary data concerning the TaBx, and HlBx genes are lacking. Generally, the ScBx genes were the most similar to the TaBx genes and the least similar to the ZmBx genes, except for ScBx1 which was the most different from HlBx1 (for more details see supplementary materials, Tables 5, 6 and 7). Polymorphisms (SNPs and INDELs) were found in all ScBx gene components. The frequency of SNPs and INDELs ranged from 0.08–5.15 and 0.0–1.11, respectively, depending on the gene and the gene component. The number of polymorphisms was usually higher in introns and 3′UTRs (up to tenfold) than in exons, with the exception of ScBx5 compared with ZmBx5 when the opposite relationship was observed. The most frequently observed SNPs in the ScBx genes were G/A, A/G, C/T and T/C, and the rarest were A/T, T/A, G/T and T/G, independent of the gene and species with which they were compared. Numerous insertions and deletions were identified in the ScBx genes, usually in non-coding components, especially compared to the ZmBx and HlBx sequences. The length of some detected INDELs reached nearly 1000 bp. The longest continuous insertion (1006 bp) was present in the 3′UTR of KF-ScBx5 gene compared with HlBx5 and the longest deletion (166 bp), in the 3′UTR of KF-ScBx4 gene compared with ZmBx4 (for more details see supplementary materials, Table 7).
When the coding sequences of Bx genes were compared, the highest numbers of polymorphisms were found in the first and second exons of ScBx1, and the first and the third exons of ScBx2 ÷ ScBx5. The second exon of the cytochrome P450 monooxygenase coding genes (Bx2 ÷ Bx5) was the least polymorphic in all species.
In general, rye Bx genes are most structurally similar to TaBx genes and then to HlBx genes. However, relatively extensive differences at the nucleotide level, particularly in the introns, may cause variable splicing in these species. At least some of the SNP-connected conservative AA substitutions and/or INDEL-connected AA insertions/deletions are likely to influence the properties of the encoded proteins.
Phylogenetic analysis of Bx genes
Bx1
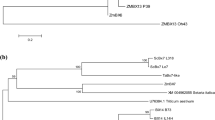
A phylogenetic tree constructed for the Bx1 “branchpoint” gene of Secale cereale, Triticum aestivum, Hordeum lechleri and Zea mays showed four clusters corresponding to the compared species except for TaBx1 from B genome which was located together with ScBx1 genes (Fig. 2). The KF-ScBx1 gene was the closest neighbor of HG-ScBx1. Among the wheat genes, the TaBx1 from genome B appeared to be the most closely related to KF-ScBx which was confirmed by direct sequence comparison (for more details see supplementary materials, Tables 5, 6, Fig. 3). These relationships between the analyzed species support the hypothesis of Dutartre et al. (2012) that the duplication of tryptophan synthase α (TSA) and its neofunctionalization, leading to the creation of Bx1, occurred before the radiation of Poaceae and the separation of the Pooideae and Panicoideae.
Bx2 ÷ Bx5
Phylogenetic analysis of the four ScBx genes encoding cytochrome P450s demonstrated that the newly isolated KF-ScBx2 ÷ ScBx5 genes are closely related to the corresponding rye HG-ScBxs (Fig. 3). Unfortunately, their similarity to the J -ScBx3 ÷ -ScBx5 is rather not possible to determine as the data are available only for J -ScBx2. In comparison with other species, these KF-ScBx genes generally appeared to be most similar to the corresponding wheat TaBx gene. A similar relationship was revealed by La Hovary (2012). As previously shown by Frey et al. (2009), each of the genes Bx2 ÷ Bx5 were from a separate clade, which is a strong indication that the progenitors of these genes evolved before the divergence of the Triticeae and the Panicoideae.
The results of phylogenetic analysis of Bx genes from three different rye accessions, wheat, maize and H. lechleri were generally consistent with the findings of earlier studies (Frey et al. 2009; La Hovary 2012; Dutartre et al. 2012) and support the proposed co-evolution of genes controlling the biosynthesis of BXs in the Poaceae family and their monophyletic origin.
Conclusion
-
A detailed analysis of the newly isolated ScBx1 ÷ ScBx5 genes showed that they are similar to the Bx genes of other Poaceae species, both at the structural and, most probably, the functional level. However, some of the identified polymorphisms may cause slight differences in their expression specificity both on RNA and protein levels.
-
The introns and 3′UTRs of ScBx genes (particularly ScBx3 and ScBx4) characterized in this study represent gene components under strong evolutionary pressure. In spite of their regulatory role, these sequences may serve as a unique and valuable resource for genetic diversity studies or for association mapping and genomic selection.
-
The presence of stress-specific DNA regulatory motifs (especially cis-acting regulatory elements involved in the MeJA-responsiveness) in the promoters of ScBx genes indicates their significant role in the benzoxazinoid-dependent defence strategy.
Notes
C-score is a confidence score for estimating the quality of predicted models by I-TASSER. It is calculated based on the significance of threading template alignments and the convergence parameters of the structure assembly simulations.
References
Dutartre L, Hilliou F, Feyereisen R (2012) Phylogenomics of the benzoxazinoid biosynthetic patway of Poaceae: gene duplications and origin of the Bx cluster. BMC Evol Biol 12(64). doi:10.1186/1471-2148-12-64
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Frey M, Kliem R, Saedler H, Gierl A (1995) Expression of a cytochrome P450 gene family in maize. Mol Gen Genet 246(1):100–109. doi:10.1007/BF00290138
Frey M, Chomet P, Glawischnig E, Stettner C, Grün S, Winklmair A, Eisenreich W, Bacher A, Meeley RB, Briggs SP, Simcox K, Gierl A (1997) Analysis of a chemical plant defense mechanism in grasses. Science 277:696–699. doi:10.1126/science.277.5326.696
Frey M, Schullehner K, Dick R, Fiesselmann A, Gierl A (2009) Benzoxazinoid biosynthesis, a model for evolution of secondary metabolic pathways in plants. Phytochemistry 70:1645–1651. doi:10.1016/j.phytochem.2009.05.012
Grün S, Frey M, Gierl A (2005) Evolution of the indole alkaloid biosynthesis in the genus Hordeum: distribution of gramine and DIBOA and isolation of the benzoxazinoid biosynthesis genes from Hordeum lechleri. Phytochemistry 66(11):1264–1272. doi:10.1016/j.phytochem.2005.01.024
Jonczyk R, Schmidt H, Osterrieder A, Fiesselmann A, Schullehner K, Haslbeck M, Sicker D, Hofmann D, Yalpani N, Simmons C, Frey M, Gierl A (2008) Elucidation of the final reactions of DIMBOA-glucoside biosynthesis in maize: characterization of Bx6 and Bx7. Plant Physiol 146:1053–1063. doi:10.1104/pp. 107.111237
Kato-Noguchi H (2008) Effects off our benzoxazinoids on gibberellin- induced a-amylase activity in barley seeds. J Plant Physiol 165:1889–1894. doi:10.1016/j.jplph.2008.04.006
La Hovary C (2012) Allelochemicals in Secale cereale: Biosynthesis and molecular biology of benzoxazinones. http://gradworks.umi.com/34/63/3463787.html
Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y et al (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30:325–327. doi:10.1093/nar/30.1.325
Makleit P (2005) Changes in cyclic hydroxamic acid content of various rye varieties for the effect of abiotic stress. Acta Biol Szeged 49(1–2):103–104
Meihls LN, Handrick V, Glauser G, Barbier H, Kaur H, Haribal MM, Jander G (2013) Natural variation in maize aphid resistance is associated with 2, 4-dihydroxy-7-methoxy-1, 4-benzoxazin-3-one glucoside methyltransferase activity. Plant Cell 25(6):2341–2355. doi:10.1105/tpc.113.112409
Meyer SLF, Rice CP, Zasada IA (2009) DIBOA: fate in soil and effects on root – knot nematode egg numbers. Soil Biol Biochem 41:1555–1560. doi:10.1016/j.soilbio.2009.04.016
Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8(19):4321–4326. doi:10.1093/nar/8.19.4321
Nei M, Kumar S (2000) Molecular evolution and phylogenetics. Oxford University Press, New York
Niemeyer HM (2009) Hydroxamic acids derived from 2-hydroxy-2H-1,4-benzoxazin-3(4H)-one: key defense chemicals of cereals. J Agric Food Chem 57:1677–1696. doi:10.1021/jf8034034
Nomura T, Ishihara A, Imaishi H, Ohkawa H, Endo TR, Iwamura H (2003) Rearrangement of the genes for the biosynthesis of benzoxazinones in the evolution of Triticeae species. Planta 217:776–782. doi:10.1007/s00425-003-1040-5
Nomura T, Ishihara A, Yanagita RC, Endo TR, Iwamura H (2005) Three genomes differentially contribute to the biosynthesis of benzoxazinones in hexaploid wheat. PNAS 102(45):16490–16495. doi:10.1073/pnas.0505156102
Nomura T, Nasuda S, Kawaura K, Ogihara Y, Kato N, Sato F, Kojima T, Toyoda A, Iwamura H, Endo TR (2008) Structures of the three homoeologous loci of wheat benzoxazinone biosynthetic genes TaBx3 and TaBx4 and characterization of their promoter sequences. Theor Appl Genet 116:373–381. doi:10.1007/s00122-007-0675-1
Oikawa A, Ishihara A, Iwamura H (2002) Induction of HDMBOA-Glc accumulation and DIMBOA-Glc4-O-methyltransferase by jasmonic acid in poaceous plants. Phytochemistry 61:331–337. doi:10.1016/S0031-9422(02)00225-X
Peterson DG, Tomkins JP, Frisch DA, Wing RA, Paterson AH (2000) Construction of plant bacterial artificial chromosome (BAC) libraries: an illustrated guide. J Agric Genomics 5:1–100
Roy A, Kucukural A, Zhang Y (2010) I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc 5:725–738. doi:10.1038/nprot.2010.5
Šimková H, Číhalíková J, Vrána J, Lysák MA, Doležel J (2003) Preparation of HMW DNA from plant nuclei and chromosomes isolated from root tips. Biol Plant 46:369–373. doi:10.1023/A:1024322001786
Šimková H, Rakoczy-Trojanowska M (2010) Construction of rye (Secale cereale L.) BAC library. III-rd Polish Congress of Genetics. Lublin, Poland, September 12–15 2010:198
Sue M, Nakamura C, Nomura T (2011) Dispersed benzoxazinone gene cluster: molecular characterization and chromosomal localization of glucosyltransferase and glucosidase genes in wheat and rye. Plant Physiol 157:985–997. doi:10.1104/pp.111.182378
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. doi:10.1093/molbev/mst197
Virtanen AI, Hietala PK (1955a) 2(3)-Benzoxazolinone an anti-fusarium factor in rye seedlings. Acta Chem Scand 9:1543–1544
Virtanen AI, Hietala PK (1955b) The structure of the precursors of benzoxazolinone in rye plants. II. Suomen Kemistilehti 32:252
Wahlroos 0, Virtanen AI (1959) Precursors of 6-methoxybenzoxazolinone in maize and wheat plants, their isolation and some of their properties. Acta Chem Scand 13(9):1906–1908. doi:10.3891/acta.chem.scand. 13-1906
Zhang Y (2008) I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9(40). doi: 10.1186/1471-2105-9-40
Zasada IA, Rice CP, Meyer SLF (2007) Improving the use of rye (Secale cereale) for nematode management: potential to select cultivars based on Meloidogyne incognita host status and benzoxazinoid content. Nematology 9:53–60. doi:10.1163/156854107779969745
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by: Andrzej Górny
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOC 638 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Bakera, B., Makowska, B., Groszyk, J. et al. Structural characteristics of ScBx genes controlling the biosynthesis of hydroxamic acids in rye (Secale cereale L.). J Appl Genetics 56, 287–298 (2015). https://doi.org/10.1007/s13353-015-0271-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13353-015-0271-z