Abstract
The critical care environment drives huge volumes of data, and clinicians are tasked with quickly processing this data and responding to it urgently. The neurocritical care environment increasingly involves EEG, multimodal intracranial monitoring, and complex imaging which preclude comprehensive human synthesis, and requires new concepts to integrate data into clinical care. By definition, Big Data is data that cannot be handled using traditional infrastructures and is characterized by the volume, variety, velocity, and variability of the data being produced. Big Data in the neurocritical care unit requires rethinking of data storage infrastructures and the development of tools and analytics to drive advancements in the field. Preprocessing, feature extraction, statistical inference, and analytic tools are required in order to achieve the primary goals of Big Data for clinical use: description, prediction, and prescription. Barriers to its use at bedside include a lack of infrastructure development within the healthcare industry, lack of standardization of data inputs, and ultimately existential and scientific concerns about the outputs that result from the use of tools such as artificial intelligence. However, as implied by the fundamental theorem of biomedical informatics, physicians remain central to the development and utility of Big Data to improve patient care.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The entire digital universe in 2020 consists of 40 times more bytes than there are stars in the sky—44 zettabytes. In just a few years, we will collectively create the equivalent of nearly 9 million DVDs worth of data every hour [1]. More than 90% of the data in the world has been created in just the past several years [2]. Healthcare data is also accumulating rapidly with expanding adoption of electronic health record (EHR) systems, and what was projected to have surpassed 25 exabytes just a few short years ago [3] is now estimated to balloon to 35 zettabytes by 2020 [4]. The critical care environment, in particular, drives huge volumes of data from a variety of sources, everything from intermittent imaging results to continuous physiological sensor data. Clinicians are tasked with processing this data and responding to it urgently in order to care for the sickest patients. As brain-focused critical care grows internationally, the use of data-intensive brain monitoring technologies such as electroencephalography (EEG) and intracranial multimodal monitoring (MMM) generates time-resolute multivariate time series that preclude comprehensive human interpretation and require new concepts to integrate these data into patient care and to leverage these data to foster research insights and progress.
What Is Big Data: Defining Big Data in Neurocritical Care
In 2012, the Big Data Research and Development Initiative was launched by the USA, leading the National Institute of Standards and Technology (NIST) to create a Big Data Interoperability Framework including definitions to guide concepts surrounding large-volume data in industries such as healthcare. Big Data is a term that encompasses any data that cannot be handled efficiently by “traditional data architectures,” such as relational databases (e.g., standardized query language (SQL)) or consumer software platforms (e.g., Microsoft Excel) [5]. The canonical 4 Vs of Big Data—volume, variety, velocity, and variability—describe characteristics of data that make it a challenge to handle with standard technologies. Big Data by definition requires “a scalable architecture for efficient storage, manipulation, and analysis” [5].
The critical care environment generates data every day that embody the characteristics (Vs) of Big Data. In the pediatric intensive care unit (ICU), one study documented a median of 1348 clinical data points each day, with nearly double the volume in patients requiring the most advanced technologies such as extracorporeal membrane oxygenation [6]. ICU data comes from a variety of systems with multiple standards, including structured data (Health Level Seven (HL7) clinical flowsheet data), propriety-format semi-structured sensor data such as EEG, and unstructured free-text clinician notes, as well as Digital Imaging and Communications in Medicine (DICOM) standard imaging and Logical Observation Identifiers Names and Codes (LOINC) standard laboratory values. While there are some technical standards, meta-data—such as the zeroing strategy used to measure the arterial blood pressure, a key variable in interpreting cerebral perfusion pressure—is lacking and there are no widely adopted methods for integrating these data [7]. The high-velocity flow of this data occurs in real time, and clinicians in the ICU are expected to log in to different systems in order to respond to raw data as quickly as possible. Clinicians have to account for the variability in each data point over time. For instance, patients undergoing thrombectomy after stroke in the USA require blood pressure measurements each 15 min initially; these data points may be validated by nursing staff from an arterial blood pressure catheter that delivers second-by-second data. Within a single day, these recorded values are spaced to every hour and become sourced from noninvasive blood pressure cuff measurements performed directly by a bedside nurse.
Additional Big Data Vs have been proposed with direct implications on critical care data (Table 1). The veracity of the data in the ICU is constantly evaluated: clinicians may ask for the external ventricular drain to be re-zeroed or frequently ignore a concerning but artifactual rhythm on telemetry that results from sternal rub. Validity and value are related terms that are similarly considered as part of day-to-day care. In the recent Brain Oxygen Optimization in Severe Traumatic Brain Injury (BOOST)-II trial, the first 5 h of brain tissue oxygen data was excluded from analysis to allow for equilibration and validity checks [8] and a median of 8 h was required for valid brain tissue oxygen data in another retrospective series [9]. The value of data is equally important. For instance, a lab test such as a brain natriuretic peptide level in a patient with neurocardiogenic stunning and pulmonary edema may add nothing to the clinical exam in a patient with known extravascular volume overload. In contrast, several hours of continuous EEG monitoring in a comatose patient with nonspecific background slowing may continue to have value based on evidence that a full 48 h of EEG may be required for adequate sensitivity to detect relevant pathology, such as seizures [10].
Big Data Infrastructure in Neurocritical Care
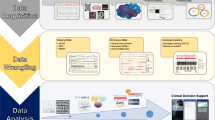
Informatics relies on a simple model wherein input is processed and analyzed to produce output (Fig. 1). However, when the input consists of Big Data, new types of architecture are required by definition. A key step in creating a Big Data infrastructure from an engineering standpoint is to move away from traditional relational database systems. Termed NoSQL databases, these are infrastructures that allow flexible data storage within a distributed network of computing resources. Whereas SQL databases use tables to store data, which include definitions about which data is stored where, NoSQL databases can be stored free-form or can use novel strategies based on key values or graph networks. In many instances, developing the architecture necessary to store and use Big Data captured in the ICU requires collaboration with local resources in order to reduce costs and comply with information security and technological regulations.
A simplified Big Data Framework. Informatics relies on input to generate an output. Input is made up of data from a variety of variable sources. Preprocessing (cleaning, artifact removal, and signal processing) steps are performed so that the next steps reflect only valid, verified data. Data is simplified through feature extraction, and then statistical processing is used to evaluate the data for meaning and importance, whether through standard statistical inference techniques or more sophisticated artificial intelligence algorithms. Finally, analytics are used to describe the data, then use the data to predict relevant events or to prescribe specific courses of action that might be warranted based on the data.
Tools then must be developed for preprocessing, feature extraction, and statistical processing (Fig. 1). Preprocessing may refer to outlier detection algorithms that remove values of arterial pressure that are negative in value, complex algorithms designed to check data based on its relationship with other data (e.g., two temperatures from two sources may be checked to see if one is disparate), or imputation procedures to fill in the blanks where data might be missing. Feature extraction subsequently reduces the raw data into discrete data points while allowing for data integration through common sampling frequencies (e.g., daily averages). Statistical processing refers to methods of evaluating the data either using statistical inference or using artificial intelligence tools. Finally, analytics refer to the output product depending on what is needed: description, prediction, or prescription.
Clinicians are intuitive data scientists. Consider a patient who has tachycardia. First, the ECG telemetry signal itself is evaluated for accuracy (preprocessing) and the heart rate, which is a 10-s average, is then recorded in a flowsheet with other down-sampled hourly heart rate values (feature extraction) by a bedside nurse. The clinical care team then performs time series analysis with Bayesian inference (statistical processing) to decide that this value is higher than expected based on prior values even though a value or 2 was not recorded overnight. An internal model is created incorporating information from EHR including a) that the patient has had a Foley catheter for several days and b) she has new leukocytosis, and with that, the care team is now able to predict that the patient is developing sepsis (analytics) and sends a urinalysis to confirm rather than waiting for a catastrophic change in physiology to declare itself. All this is done without a second thought: it is how we are trained to think. But adding the complexity of the critical care unit to this problem – the patient also had a venous thrombosis found last night and is being weaned from her analgosedation – and it is easy to see how quickly we are subjected to thousands of data points prior to arriving at an analysis. Despite our abilities, humans are limited in consciously processing more than four independent variables at the same time [11].
Insights Through Feature Extraction
Much of what is referred to as Big Data in the acute setting results from high-volume time series data, which requires signal processing in order to both preprocess and extract relevant features. Common signal preprocessing techniques include filtering based on specific frequencies or outlier removal methods (Fig. 2). Signals such as the intracranial pressure (ICP) waveform are often expressed using summary statistics as a principle feature, such as mean ICP. However, clinicians recognize each wave within the signal has physiologic importance. An example of detailed signal processing in neurocritical care is illustrated by the Morphological Clustering and Analysis of ICP (MOCAIP) framework [12], which was developed to precisely measure nonartifactual ICP peaks, thereby creating a rich feature set including automated measurements of slope and waveform duration for each beat-to-beat ICP oscillation.
Common signal processing steps. Signal processing techniques are used for raw sensor data in order to remove artifacts and noise that may impact feature extraction and therefore analysis. (A) The raw signals recorded in the ICU are often noisy. The ICP waveform recorded here is visible, but there is high-frequency data that gives the signal a fuzzy appearance. Electrical noise (60 Hz in the USA or 50 Hz in Europe) is common. (B) A filter is applied that removes frequencies of 60 Hz from the signal; note the fuzziness of the ICP waveform is gone. (C) Simple outlier detection uses simple thresholds to dismiss any signal outside of that threshold. For instance, an ICP > 100 mmHg is not likely to be physiologic and, therefore, any values outside of this area can be excluded. Similarly, (D) the oscillations within a signal can be used as thresholds by stipulating that a signal should have a certain standard deviation over a window of n units of time. In this case, there were 3 s of flat, invariant values suggesting an artifact that should be removed.
Physiologic time series vary according to complex interactions that are not explained by simple assumptions (sometimes referred to as nonlinear relationships). The implication is that a whole system cannot be understood simply by component signals. Measures of complexity uncover insight embedded within these signals that are not visible on bedside monitors. Entropy measures, for example, quantify the unpredictability or randomness within a signal; several different entropy measures have been used including approximate entropy (ApEn), sample entropy (SampEn), and multiscale entropy (MSE). In a cohort of 290 patients with severe traumatic brain injury (TBI), the MSE of the ICP signal demonstrated decreased complexity during critically elevated ICP and the average complexity index of the ICP signal was an independent predictor of outcome [13]. Scalar features are those that do not change whether the feature is measured over seconds or hours or even days which can be quantified using detrended fluctuation analysis (DFA). In one study of 147 patients with TBI, the DFA intercept and scaling exponent were linearly correlated and independently associated with outcome [14]. While measures of heart rate variability have been studied for decades, higher-order measures of complexity have only begun to touch on the signals measured in the neurocritical care unit.
The relationships between signal features can provide insight into physiologic states. In one of the most successful translational informatics stories in neurocritical care, the correlation coefficients between the input pressure (either arterial or cerebral perfusion pressure) and various surrogates of cerebral blood flow or volume (i.e., ICP or noninvasive infrared spectroscopy) have been validated as surrogate measures of autoregulatory function. The pressure reactivity index (PRx) specifically is defined as the moving average correlation coefficient of the mean arterial pressure and the ICP. In a seminal series of 327 patients with severe TBI undergoing ICP monitoring, the PRx was mapped to cerebral perfusion pressure (CPP), uncovering an optimum CPP at which autoregulatory function was maximized. The distance from this optimum CPP was shown to vary linearly with outcome [15], reinforced by a meta-analysis demonstrating that particularly in TBI, there is a significant correlation (r = 0.37; 95% CI, 0.28–0.47) with functional outcome [16]. Coupling between more than two variables becomes more complex because the number of correlations increases exponentially with each new variable. Work to develop a framework for quantifying the nonlinear interactions between networks of multiple physiologic variables has led to the concept of time delay stability which has been studied in the context of sleep-state transitions and promises to uncover novel cross-system coupling associated with pathological brain states [17]. Graph theoretical models allow quantification of networks of physiological interactions based on correlations or more complex nonlinear interactions, while techniques such as Granger causality may play a role in determining the direction of those relationships.
For unstructured data such as free-text notes, natural language processing (NLP) is a tool that draws features from syntactical data based on specified rules. Features can then be generated directly from clinical documentation, potentially preserving the nuances of uncertainty and allowing for data that has no structured access point. NLP has been used primarily to enhance research and registry data collection, including use in abstracting key data from CT reports about the size and location of subdural hematomas [18], identifying cohorts of patients with subarachnoid hemorrhage (SAH) and their matched controls [19], and recognizing ischemic stroke subtypes based on progress notes and radiology reports [20].
Insights Through Artificial Intelligence
Statistical processing tools refer to both traditional statistical inference models, such as linear or binary regression, or artificial intelligence tools, including machine learning and neural network techniques. Regression models have driven much of the statistical methodology of the past decades; however, there are significant limitations on the number of predictor variables a model may contain and they may make assumptions about the relationships between those predictors that are not true, e.g., assumptions about linearity between variables. As a general principle, the “one in ten” rule dictates that at least 10 observations should be made for each predictor. Thus, a small study of 100 patients might include 10 predictors for a linear endpoint; however, if the model is designed to predict death and only 20 patients died, at most 2 predictors should be used. Modern alternatives to standard binary regression analysis techniques may relax this rule, but in general, this rule provides reasonable guidance [21].
Machine learning tools, on the other hand, do not require these restrictions a priori. Instead, overfitting becomes the primary limitation. Overfitting refers to the development of a complex model that works on the small n used to train the model, but because the model is so specific, it is inaccurate when applied to larger test datasets with higher variance. This generalization gap is a reflection of the bias–variance trade-off (Fig. 3). Bias refers to a model that does not incorporate fully the complexity of the data whereas variance reflects noise from data that is not needed to create a valid model. Therefore, machine learning often seeks to find the model with the lowest prediction error by varying parameters. Machine learning can model known endpoints, termed supervised machine learning, or it can explore relationships within the data in a hypothesis-free way, referred to as unsupervised machine learning. A deeper look at the various machine learning tools that are readily available is covered elsewhere [22].
The bias–variance trade-off. This figure conceptually demonstrates the trade-off between bias (red line), meaning that data is not included in the model that is necessary for the model to be accurate, and variance (green line), or data that is not necessary for the model to be accurate, sometimes referred to as noise. The model error is the sum of the bias2, the variance, and irreducible error, as shown in the bottom portion of the figure. The Y-axis represents prediction error, and the X-axis represents the complexity of the model being created. Training error (dashed orange line) refers to increasing accuracy when variables are added to a model being created on relatively small cohort. However, this model may be overfit and this leads to an increase in test or validity error (dashed blue line) when applied to larger cohorts. An optimal model falls on the dashed blue line at its lowest, when bias and variance are mutually minimized, as denoted as the relatively white space between the underfitting and overfitting zones demonstrated in gray shading. Across the top are visualizations of models that are representative of an underfit model on the left (such as a linear regression, which fails to capture the complexity of the data) and an overfit model on the right, in which the model adheres to the training data that it would be inaccurate given new data points. An optimal fit is shown in the center, in which an exponential curve is fit nicely across the data in a way that minimizes the distance between the observed and the predicted data.
Supervised Learning
Supervised learning can be used for high-dimensional datasets in order to model or describe how the data relates to a label. In a landmark study assessing conscious awareness in patients with acute brain injuries, continuous EEG features were used to construct a support vector machine model to describe responses to verbal commands (the label) using a block design. Fifteen percent of patients clinically in coma had EEGs that predicted the label with an area under the curve (AUC) significantly more than 50% (corresponding to chance alone), suggesting their EEG was changing in response to verbal commands even if their bodies were not [23].
Endpoints or outcome measures may also act as labels for supervised machine learning. Using the vital signs from nearly 270,000 hospitalized patients, one study used several machine learning techniques to predict death, cardiac arrest, or ICU admission. Using derivation and validation cohorts including more than 16,000 events, the authors found that the random forest method yielded the highest AUC of 80% compared to established models (such as the modified early warning score) [24]. Standard regression models have been validated to predict clinical outcome following moderate-to-severe TBI based on more than 15,000 patients enrolled in two large clinical trials using simple admission variables [25]. The Brain Monitoring with Information Technology (BrainIT) group used Gaussian process models in a cohort of 264 patients with TBI and demonstrated that the inclusion of ICP and arterial blood pressure signals sampled each minute over 4-h windows across 239 patients increased the AUC of the International Mission for Prognosis and Analysis of Clinical Trials (IMPACT) model for long-term outcome from 72 to 90% [26].
Importantly, caution is warranted prior to deploying complex machine learning algorithms for outcome prediction: as practice and patients change in their baseline characteristics, models need not only validation, calibration, and decision curve analyses [27], but model versioning and regular updating in order to avoid applying improperly fit or out-of-date models. In other words, machine learning models act more like software than statistics; in this regard, regression methods will likely continue to play an important role in prognostic modeling [28].
Unsupervised Learning
Unsupervised techniques do not require labels or an outcome endpoint but rather group input data based on patterns that do not need to be defined a priori, thus reducing some of the risk of investigator bias. For instance, in a cohort of pediatric patients with sepsis, the expression of thousands of genes was examined using unsupervised techniques, and despite the individual variability of gene expression across each patient, there were essentially three distinct “clusters” or patterns that emerged [29]. By focusing on these clusters, the authors found that they differed based on adaptive immunity and zinc biology–related gene pathways and subsequently confirmed they also differed in their clinical characteristics. Grouping patients first by age or severity and using a supervised technique might have missed these insights: groups B and C had similar illness severity, and groups A and C had similar ages.
Unsupervised techniques include principal component analysis used for dimensionality reduction, in which the structure of the data itself is simplified into representative features. Simplified data structures may have stand-alone importance when it defines a class or state in which a patient exists by organizing multivariate data into groups, termed cluster or latent class analysis. In one study exploring hierarchical cluster analysis in 23 patients with TBI, 18 brain and body signals were measured each minute for a median of 7 days from which three clusters emerged, roughly corresponding to outcome [30]. In a follow-up from the same group, 17 patients were studied each with 45 measured signals; 10 clusters were identified. Assessment of these clustered patients by clinicians failed to identify any clinical correlation, yet several clusters were associated with a higher probability of death or multiorgan failure [31]. Interestingly, cluster assignments shifted over time, suggesting that clustering algorithms may identify novel hidden physiologic states that might be potentially influenced in order to impact outcome.
The presence of hidden states within time series physiologic data also forms the rationale for using unsupervised techniques as a feature generation tool in predictive modeling. In a study of 488 patients with subarachnoid hemorrhage, a convolution dictionary learning algorithm was used to generate 4200 physiologic features. When these features (similar to very small clusters or states) were combined with clinical variables in a partial least squares regression, the model resulted in an AUC of 78% (95% CI, 64–92) [32].
Topological data analysis is an unsupervised technique that includes dimensionality reduction, pattern detection, and network visualization and allows for an assessment of complex interrelationships within multidimensional data. In a proof-of-concept study using preclinical TBI and spinal cord injury models, novel relationships between blood pressure and outcome and a robust interaction between brain and spinal cord injuries were found, despite the fact that the data had been studied rigorously using standard statistical techniques [33]. Future work using topological data analytic tools in large observational, multidimensional clinical datasets such as the Transforming Research and Clinical Knowledge in Traumatic Brain Injury (TRACK-TBI) study holds promise to undercover hypothesis-generating relationships between clinical, physiologic, and radiologic data.
Learning algorithms are combined approaches that rely on unsupervised feature extraction followed by interactive expert annotation and act as a method of making expensive or time-intensive labeling more efficient. In a study of ICP waveform identification and artifact reject, 34 patients with ICP waveform data were used to generate more than 125,000 dominant pulse waves. An active learning paradigm was then employed to select specific waveforms for expert labeling in order to maximize the Fisher information or minimize variance. Active learning is a technique by which the algorithm attempts to learn by asking the user to provide a label (“is this an artifact or a true ICP waveform?”) and then uses those labels to iteratively improve its modeling. By doing it so, active learning reduces the amount of labels necessary for the algorithm to learn. Thus, as few as 50 labeled ICP segments were required per rater, resulting in AUCs averaging 95%, far above template matching and threshold-based comparisons [34]. In a similar paradigm leveraging interactive expert labeling, continuous EEG signals were clustered in order to provide efficient annotation of seizures or abnormal periodic or rhythmic discharges. A total of 369 continuous EEG recordings were broken into segments based on changes in background features that were then described using bag-of-words representation with each cluster representing a different background pattern. This allowed expert readers to quickly scan 30 to 50 clusters and identify patterns, quantifying 24 h of EEG in a mere 3 min relative to reading and annotating each window across the entire recording [35].
Hidden Markov Models
Hidden Markov models are dynamic Bayesian networks in which hidden states occur with a probability that is based on the existing state of the observed data. Bayesian inference leverages existing data to model what we expect (prior probability) and new data to model what we see (likelihood), generating a posterior probability, which takes into account both expected and existing data. Markov processes are those in which the probability of a certain state depends on the prior state; the term “hidden” refers to the fact that these states are not observable or defined. Therefore, the hidden Markov model uses posterior probabilities to identify states that might not be directly observable. In a recent cohort of patients with TBI and detailed physiologic recordings, nearly 29,000 h of data across 379 patients was explored using a hidden Markov model. States were based on a combination of cerebral perfusion pressure, ICP, pressure reactivity, and compensatory reserve. Three distinct states were identified, and the model was able to predict death based on these states with an AUC of 0.78 ± 0.07. States were dose-dependent, in which the proportion of time spent in the “worst” state also predicted death with an AUC of 0.75 ± 0.07 [36]. Importantly, the average ICP in the “worst” state was 20.8 mmHg, less than the recommended threshold to consider treatment based on the most recent Brain Trauma Foundation guidelines [37]. Similarly, using a combination of hidden Markov modeling and statistical inference, an autoregressive ordinal regression technique was used to predict critical elevations in ICP across 817 patients and critically low brain tissue oxygen across 242 patients. Using this method, they found that a “pre-crisis” ICP of just 19 mmHg predicted ICP crisis within the next 30 min with a cross-validation AUC of 86% [38].
Neural Networks
Neural network algorithms use hidden layers of nodes, or neurons, to create a hierarchy of features that better represent the nonlinear structure of input data. Artificial neural networks and convolutional neural networks can be relatively simple (e.g., one or a few layers) or can encompass multiple layers, so-called deep learning. Deep learning, in particular, has been leveraged for imaging data, including identification and quantification of stroke or hemorrhage burden [39]. Even the apparent age of the brain can be intuited using deep learning strategies, a promising method of inferring premorbid health and cognitive function. Convolutional neural networks were used across 2001 healthy controls to derive brain age which matched actual age with a correlation coefficient of 0.96 and a mean absolute error of only ~ 4 years [40]. Neural networks may be more capable of handling the complexity of data generated in the ICU setting. For instance, neural networks outperform linear regression and support vector machine techniques more than 90% of the time for prediction of clinically relevant outcomes across a variety of neurosurgical applications [41]. However, moving from intuitive linear relationships to accepting a black box of hidden nodes is a challenge for most bedside clinicians.
Integrating Big Data into Research
Until Big Data goes from in silico to in vivo, much of the potential patient impact that is promised by Big Data analytics will continue to be developed and refined post hoc from data that comes from increasingly large, well-curated, and more complete datasets. From time series physiologic data to genome-wide linkage associations, large sets of data with rich clinical phenotyping will be necessary to reduce bias and ensure Big Data tools will be powered for population-level insights while accurate enough for eventual use at the individual level. Efforts to curate large, complex datasets have been initiated in the international TBI community and are beginning to bear fruit; for example, the International Initiative for TBI Research (InTBIR) [42] acts as a framework for international collaboration by providing a way for large, completed funded studies to be aggregated, greatly expanding knowledge generalizability.
Despite the primacy of the randomized controlled trial (RCT) in modern medicine, there is a growing conviction that the design of such trials undermines the efficacy of therapies applied without precisely targeting the right patients for the therapy [43]. The concept of precision medicine was pioneered through the genetic analysis of tumors and the development of tumor-specific chemotherapies, a thoroughly Big Data approach that has continued to evolve. Modern approaches to neurocritical care research should leverage the strengths of Big Data and the randomized controlled trial design [43].
Consider a hypothetical RCT of a candidate neuroprotective agent to prevent delayed cerebral ischemia after SAH: candidate single nucleotide polymorphisms are identified [44] and added to imaging-based risk models [45] on admission to stratify patients into groups. Real-time physiologic data such as heart rate or blood pressure are then used to adjust this risk profile over time [32]. As patients enter the window for delayed cerebral ischemia, those with the highest risk could then be randomized to various agents using an intermediate biomarker such as the development of spreading depolarizations [46] as an endpoint; agents that do not perform well are gradually dropped from random assignment. By working in a multidisciplinary way to integrate genetics, neurocritical care, data science, neurophysiology, and adaptive clinical trial design, the most promising interventions can be evaluated in a targeted population most likely to benefit at a much lower cost.
Increasingly, there is need to evaluate the evidence behind critical care in general. For instance, physiology is constantly manipulated to achieve goals that have little evidence-based support. Instead of testing a drug, Big Data–derived computational analytics can be used to drive precise physiologic targets. By using modular, app-based design, data scientists can facilely move post hoc algorithms into the clinical space to prospectively test specific critical care management strategies. For instance, an algorithm to identify an optimum CPP from the PRx was developed as a visual interface (via ICM+; Cambridge, UK) capable of being deployed within systems without homegrown Big Data infrastructure. This algorithm is now being used to guide the management of CPP in patients with severe TBI as part of the COGITATE study (NCT02982122) [47]. While the US Food and Drug Administration (FDA) does not require specific approval for Medical Device Data Systems (MDDS), defined as hardware or software intended to “transfer, store, covert formats, and display medical device data or medical imaging data” (Section 520(o)(1)(D)), analytic tools that modify the ICP and CPP data or its display may require regulatory approval. Therefore, this paradigm could lead to FDA approval of the data tools that guide therapy, much in the way that the FDA approves and regulates tested biologic agents currently.
These examples leverage both Big Data and randomized controlled trial design in order to address a lack of evidence within neurocritical care by maximally enriching the patient cohorts that stand to benefit the most from a therapeutic agent or physiologic manipulation. In contrast, a second data-driven focus of research in healthcare capitalizes on the variability in clinical practice as a method of comparative effectiveness research. The concept of the learning health system has been defined by the Institute of Medicine as a way to “generate and apply the best evidence for the collaborative health-care choices of each patient and provider, drive the process of discovery as a natural outgrowth of patient care, and ensure innovation, quality, safety, and value in healthcare” [48]. From a practical standpoint, learning health systems allow patients and their caregivers to prioritize research questions and outcome endpoints, and leverage common data elements across multiple centers via the EHR in order to track improvements in those outcomes over time. Learning health systems thus are able to flexibly answer questions that would otherwise require expensive clinical trials by using practical implementation in order to shift outcomes that are important to patients or their caregivers directly. The ImproveCareNow Network is a successful example of such an architecture in the pediatric outpatient population and demonstrates the power of such a system to change both practice and outcome [49]. To date, there is no published implementation of learning health systems in the critical care environment despite its potential for common diseases such as TBI or stroke [50].
Challenges in Implementing Big Data at the Bedside
The use of Big Data focuses on three principle goals: description, prediction, and prescription. Each of these goals have profound implications for implementation. However, there are major barriers to the widespread deployment of Big Data infrastructure and techniques within neurocritical care and there are very few successful examples of a truly data-driven system within critical care in general [22].
First, many institutions do not have an infrastructure amenable to the acquisition and storage of neurocritical care data within a nonrelational database structure. Systems such as Amazon Web Services are open source and tied to platforms with data storage capacity, yet issues surrounding privacy, security, access, and bandwidth [22, 51] must be addressed by the local institution and its information technology experts. While these platforms are built to support Health Insurance Portability and Accountability Act (HIPAA) compliance, user-level configuration may lead to unsecured data. For instance, the Patient Home Monitoring Corporation reportedly configured a repository of 47 GB of patient-level documents such that they were freely available via the Internet [52] even though the Amazon S3 storage platform housing those documents is fully HIPAA-compatible.
To study or share local data, the oversight of both the institutional review board and the institution’s legal office is required to ensure intellectual property is preserved for the investigator or the institution. Ethics questions center around privacy, informed consent, justice, trust, and data ownership [53]. This is highlighted by Google’s recent acquisition of data from the Ascension health system, encompassing the health records of millions of patients across 21 states [54]. The deal stoked significant concerns about privacy; the HIPAA requirements for the deal required Google to sign a business associate agreement, which assures the information can only be used for the benefit of the healthcare system and could not be used in conjunction with any consumer data from Google itself. However, there is no control over how the data is used once it has been deidentified, termed secondary use. Even stripped of the 18 HIPAA identifiers, the inherent nature of some data may preclude true deidentification: consider an unusual physical abnormality which, tied to a CT from a regional university, might be enough to identify a subject allowing concatenation with consumer-level data without informed consent. To date, there are no unified rules governing the privacy of our data across sectors such as healthcare and industry, creating challenges to preserve the patient–doctor relationship when working with industry partners.
Even without an infrastructure, many neurointensivists have begun the hard work of assembling local datasets that require Big Data tools. But, the ICU is a messy place and data is frequently missing, ugly, or corrupt [51]. The quantity of the data varies from patient to patient, and the quality of the data depends on equipment and its placement, setup, and maintenance. Artifacts abound—a fact long-recognized by continuous EEG experts [55] and now being addressed for other signals such as ICP or arterial blood pressure by data scientists using increasingly sophisticated methods [56]. Researchers find themselves choosing to train clean data from a few highly curated patients leading to bias [57] or, conversely, using noisy data from a large, pragmatically curated cohort of patients, which leads bad analytics (an extension of the well-known computer science dictum: garbage in, garbage out) [22]. Work to create a consensus checklist for Data Access Quality and Curation for Observational Research Designs (DAQCORD; daqcord.org) is underway and promises to improve the quality of data a priori rather than relying on post hoc data cleaning methods which can be prohibitively time-consuming.
Third, data is poorly integrated in the neurocritical care unit. In a survey of sites collecting high-resolution physiologic data as part of TRACK-TBI and Collaborative European NeuroTrauma Effectiveness Research in TBI (CENTER-TBI) studies, the biggest challenges to integration included a lack of standardization between devices and a lack of standardized labels for common measurements (e.g., brain tissue oxygen monitoring). Meta-data about devices and measurements, such as device location or sampling frequency, were rarely available [7]. Further, data recorded from different devices gets time-stamped according to different internal time clocks, leading to imperfect or impossible data synchronization. Integrated platforms, e.g., Component Neuromonitoring Systems (Moberg Solutions, Inc., Ambler, PA), BedMaster (Excel Medical, Inc., Jupiter, FL), and ICM+ (Cambridge, UK), have improved data synchronization and device labeling for many of the devices used in neurocritical care. Recently, experts working primarily with physiologic data have begun coalescing around a common file format, HDF5, which is extensible and capable of handling a variety of data sizes including both high-frequency time series and intermittent laboratory data [58]. Efforts to harmonize physiologic data between sites and studies and to develop frameworks to identify physiologic phenotypes should lead to new insights. Ultimately, linking individualized physiologic responses (i.e., the physiome) to discrete clinical events or responses to therapies is a common goal of critical care research, reflecting the individualized care intensivists provide at bedside every day.
Integrating additional information sources (e.g., radiology and genomics) with bedside physiologic and clinical data continues to be a significant challenge in part because the informatics architecture for different data types often arises out of proprietary or technical necessity, limiting generalizability. For instance, the DICOM standard for neuroimaging is used by virtually every medical imaging device to store imaging data from CT to MRI. This standard originated early after the development of CT and was designed to be comprehensive; as a result, this tag-based system is overwhelmingly complex. There are public tags that act as addresses that can be looked up in publically available DICOM dictionaries, and private tags which vendors can use for propriety information. Although the framework is universal, its implementation is far from standard and allows for distinct information to be captured in proprietary ways. Simpler formats are necessary prior to leveraging Big Data at the bedside; neuroimaging research has benefitted from a robust informatics community that has embraced the NIfTI format and provided open source tools for conversion from DICOM and propriety, legacy imaging formats [59]. Conversion pathways and increasingly flexible data formats promise to link data sources together in increasingly streamlined datasets.
Clinical annotation of verifiable ICU-based events within data captured from devices or in the EHR is yet another barrier to the use of data from the ICU. There are currently no standardized annotations for bedside events in general (e.g., bathing or receiving an urgent PRN medication), and even catastrophic events such as a medication error or cardiac arrest are variably documented in notes. The concept of “neuroworsening” is critical to define when considering using Big Data to predict important changes in the neurocritical care unit. However, neuroworsening is poorly defined in clinical practice despite its definition within federally funded TBI common data elements [60] and requires careful adjudication for use even in post hoc data analysis [61]. Nursing observations that describe a neuroworsening event might exist as free text in the EHR only, if, at all, completely separated from the data that could be used to predict such an event.
Eventually, hospitals will catch up to their corporate counterparts and Big Data infrastructures may become commonplace. Certainly, the use of predictive models as clinical decision support tools is an intuitive and pressing need. Yet, there still remains a final gap between the development and implementation of Big Data technologies and the broad clinical use of even the most rigorously developed and maintained analytical algorithms. While the use of Big Data is rapidly expanding, there is a clear danger that algorithms may be affected by (and learn from) human bias [62], and for scientists, those are many of the same biases that plague clinical trials and their translation to the bedside. As a proof of this concern, one group of researchers used the concept of the implicit association test that links the time it takes humans to pair concepts (for instance, target words such as man or woman are paired with an attribute such as career or family). Using a machine learning algorithm to identify these semantic associations within a 10-word window across a broad segment of the Internet, including some 840 billion words, they found that their algorithm replicated racial and gender bias at every turn [63].
Why is this important? Consider the hypothetical use of Big Data to develop an AI algorithm predicting outcome after cardiac arrest. Developers may pull together large datasets from clinical trials, nationwide inpatient samples, and other sources. Yet, they may neglect some key inherent clinical biases: patients enrolled in studies necessarily survived to the ICU (selection bias), ethnic disparities in withdrawal of care may mean some populations are more likely to survive (confounder), and clinicians may be more likely to provide aggressive care to a younger patient compared to an older patient, or a healthy patient compared to a patient who has struggled with prior drug overdoses (observer bias). In fact, these biases (among others) do lead to withdrawal of care decisions within 72 h of arrest in one third of those who die. However, a post hoc analysis found that an estimated 16% of those patients might have gone on to have functionally favorable survival [64] had this decision not been made. If our algorithm reflects these biases, will AI make a recommendation that leads to inappropriate withdrawal of care? And how are changes in practice, such as the use of extracorporeal cardiopulmonary resuscitation, going to impact the accuracy of recommendations made by an algorithm developed based on data from survivors who only underwent manual chest compressions? This problem arises even for regression models, such as the intracerebral hemorrhage (ICH) score, which overestimates short-term mortality in modern cohorts in part due to the fact that it was developed two decades ago [65].
Federal regulation has begun to emerge that aims to regulate how algorithms are benchmarked [66]. It is clear that developing any tool for clinical use will require careful validation, bias assessment, versioning, and updating in order to understand when and how the algorithm was created. Yet, the very real concern that physicians will lose autonomy remains. In fact, some work is likely to be displaced [57]: algorithms identifying core-perfusion mismatch were used to guide recent successful delayed embolectomy clinical trials [67, 68] largely in the absence of radiologist input. However, Big Data does not preclude or diminish clinicians. In fact, clinical implementation of Big Data requires a team of data scientists and implementation experts who are led by someone with clinical domain expertise [22]—the bedside physician.
References
Desjardins J. How Much Data is Generated Each Day? [Internet]. Apr 2019. Visual Capitalist. Available from: https://www.visualcapitalist.com/how-much-data-is-generated-each-day/. Accessed 29 Oct 2019.
Jacobson R. 2.5 quintillion bytes of data created every day. How does CPG & Retail manage it? [Internet]. Apr 2013. IBM. Available from: https://www.ibm.com/blogs/insights-on-business/consumer-products/2-5-quintillion-bytes-of-data-created-every-day-how-does-cpg-retail-manage-it/. Accessed 6 Sept 2019.
Roski J, Bo-Linn GW, Andrews TA. Creating Value In Health Care Through Big Data: Opportunities And Policy Implications. Health Affairs. 2014;33(7):1115–22.
Gui H, Zheng R, Chao M, Fan H, Xu L. An Architecture for Healthcare Big Data Management and Analysis. In: Yin X, Geller J, Li Y, Zhou R, Wang H, Zhang Y, eds. Health Information Science: 5th International Conference on Health Information Science. Springer International Publishing; 2016 154–160.
Chang WL. NIST Big Data Interoperability Framework: Volume 1, Definitions. Report No. Special Publication (NIST SP)-1500–1. Oct 2015. Available from: https://nvlpubs.nist.gov/nistpubs/SpecialPublications/NIST.SP.1500-1.pdf. Accessed 6 Sept 2019.
Manor-Shulman O, Beyene J, Frndova H, Parshuram CS. Quantifying the volume of documented clinical information in critical illness. Journal of Critical Care. 2008;23(2):245–50.
Rodriguez A, Smielewski P, Rosenthal E, Moberg D. Medical Device Connectivity Challenges Outline the Technical Requirements and Standards For Promoting Big Data Research and Personalized Medicine in Neurocritical Care. Military Medicine. 2018;183(suppl_1):99–104.
Okonkwo DO, Shutter LA, Moore C, Temkin NR, Puccio AM, Madden CJ, et al. Brain Oxygen Optimization in Severe Traumatic Brain Injury Phase-II: A Phase II Randomized Trial. Crit Care Med. 2017;45(11):1907–14.
Foreman B, Ngwenya LB, Stoddard E, Hinzman JM, Andaluz N, Hartings JA. Safety and Reliability of Bedside, Single Burr Hole Technique for Intracranial Multimodality Monitoring in Severe Traumatic Brain Injury. Neurocrit Care. 2018 Dec 1;29(3):469–80.
Claassen J, Mayer SA, Kowalski RG, Emerson RG, Hirsch LJ. Detection of electrographic seizures with continuous EEG monitoring in critically ill patients. Neurology. 2004;62(10):1743–8.
Halford GS, Baker R, McCredden JE, Bain JD. How Many Variables Can Humans Process? Psychological Science. 2005;16(1):70–6.
Hu X, Xu P, Scalzo F, Vespa P, Bergsneider M. Morphological clustering and analysis of continuous intracranial pressure. IEEE Trans Biomed Eng. 2009;56(3):696–705.
Lu C-W, Czosnyka M, Shieh J-S, Smielewska A, Pickard JD, Smielewski P. Complexity of intracranial pressure correlates with outcome after traumatic brain injury. Brain. 2012;135(Pt 8):2399–408.
Burr RL, Kirkness CJ, Mitchell PH. Detrended Fluctuation Analysis of Intracranial Pressure Predicts Outcome Following Traumatic Brain Injury. IEEE Transactions on Biomedical Engineering. 2008;55(11):2509–18.
Aries MJH, Czosnyka M, Budohoski KP, Steiner LA, Lavinio A, Kolias AG, et al. Continuous determination of optimal cerebral perfusion pressure in traumatic brain injury. Critical Care Medicine. 2012;40(8):2456–63.
Rivera-Lara L, Zorrilla-Vaca A, Geocadin R, Ziai W, Healy R, Thompson R, et al. Predictors of Outcome With Cerebral Autoregulation Monitoring: A Systematic Review and Meta-Analysis. Critical Care Medicine. 2017;45(4):695–704.
Bartsch RP, Liu KKL, Bashan A, Ivanov PCh. Network Physiology: How Organ Systems Dynamically Interact. Perc M, editor. PLOS ONE. 2015;10(11):e0142143.
Pruitt P, Naidech A, Van Ornam J, Borczuk P, Thompson W. A natural language processing algorithm to extract characteristics of subdural hematoma from head CT reports. Emergency Radiology. 2019;26(3):301–6.
Castro VM, Dligach D, Finan S, Yu S, Can A, Abd-El-Barr M, et al. Large-scale identification of patients with cerebral aneurysms using natural language processing. Neurology. 2017;88(2):164–8.
Garg R, Oh E, Naidech A, Kording K, Prabhakaran S. Automating Ischemic Stroke Subtype Classification Using Machine Learning and Natural Language Processing. Journal of Stroke and Cerebrovascular Diseases. 2019;28(7):2045–51.
Pavlou M, Ambler G, Seaman SR, Guttmann O, Elliott P, King M, et al. How to develop a more accurate risk prediction model when there are few events. BMJ. 2015 Aug 11;351:h3868.
Sanchez-Pinto LN, Luo Y, Churpek MM. Big Data and Data Science in Critical Care. Chest. 2018;154(5):1239–48.
Claassen J, Doyle K, Matory A, Couch C, Burger KM, Velazquez A, et al. Detection of Brain Activation in Unresponsive Patients with Acute Brain Injury. New England Journal of Medicine. 2019;380(26):2497–505.
Churpek MM, Yuen TC, Winslow C, Meltzer DO, Kattan MW, Edelson DP. Multicenter Comparison of Machine Learning Methods and Conventional Regression for Predicting Clinical Deterioration on the Wards. Critical Care Medicine. 2016;44(2):368–74.
Steyerberg EW, Mushkudiani N, Perel P, Butcher I, Lu J, McHugh GS, et al. Predicting outcome after traumatic brain injury: development and international validation of prognostic scores based on admission characteristics. PLoS Med. 2008;5(8):e165; discussion e165.
Güiza F, Depreitere B, Piper I, Van den Berghe G, Meyfroidt G. Novel methods to predict increased intracranial pressure during intensive care and long-term neurologic outcome after traumatic brain injury: development and validation in a multicenter dataset. Crit Care Med. 2013;41(2):554–64.
Flechet M, Grandas FG, Meyfroidt G. Informatics in neurocritical care: new ideas for Big Data. Current Opinion in Critical Care. 2016 Apr 1;22(2):87–93.
Steyerberg EW, van der Ploeg T, Van Calster B. Risk prediction with machine learning and regression methods: Risk prediction with machine learning and regression methods. Biometrical Journal. 2014;56(4):601–6.
Wong HR, Cvijanovich N, Lin R, Allen GL, Thomas NJ, Willson DF, et al. Identification of pediatric septic shock subclasses based on genome-wide expression profiling. BMC medicine. 2009 Dec 1;7(1):34.
Sorani MD, Hemphill JC, Morabito D, Rosenthal G, Manley GT. New Approaches to Physiological Informatics in Neurocritical Care. Neurocritical Care. 2007;7(1):45–52.
Cohen MJ, Grossman AD, Morabito D, Knudson MM, Butte AJ, Manley GT. Identification of complex metabolic states in critically injured patients using bioinformatic cluster analysis. Critical Care. 2010;14(1):R10.
Megjhani M, Alkhachroum A, Terilli K, Ford J, Rubinos C, Kromm J, et al. An active learning framework for enhancing identification of non-artifactual intracranial pressure waveforms. Physiological Measurement. 2019 Jan;40(1):015002.
Nielson JL, Paquette J, Liu AW, Guandique CF, Tovar CA, Inoue T, et al. Topological data analysis for discovery in preclinical spinal cord injury and traumatic brain injury. Nature Communications. 2015;6(1):8581.
Megjhani M, Alkhachroum A, Terilli K, Ford J, Rubinos C, Kromm J, et al. An active learning framework for enhancing identification of non-artifactual intracranial pressure waveforms. Physiological Measurement. 2019;40(1):015002.
Jing J, d’Angremont E, Zafar S, Rosenthal ES, Tabaeizadeh M, Ebrahim S, et al. Rapid Annotation of Seizures and Interictal-ictal Continuum EEG Patterns. Conf Proc IEEE Eng Med Biol Soc. 2018;2018:3394–7.
Asgari S, Adams H, Kasprowicz M, Czosnyka M, Smielewski P, Ercole A. Feasibility of Hidden Markov Models for the Description of Time-Varying Physiologic State After Severe Traumatic Brain Injury. Crit Care Med. 2019;47(11):e880–5.
Carney N, Totten AM, O’Reilly C, Ullman JS, Hawryluk GWJ, Bell MJ, et al. Guidelines for the Management of Severe Traumatic Brain Injury, Fourth Edition. Neurosurgery. 2017;80(1):6–15.
Myers RB, Lazaridis C, Jermaine CM, Robertson CS, Rusin CG. Predicting Intracranial Pressure and Brain Tissue Oxygen Crises in Patients With Severe Traumatic Brain Injury. Critical Care Medicine. 2016;44(9):1754–61.
Zaharchuk G, Gong E, Wintermark M, Rubin D, Langlotz CP. Deep Learning in Neuroradiology. American Journal of Neuroradiology. 2018;39(10):1776–84.
Cole JH, Poudel RPK, Tsagkrasoulis D, Caan MWA, Steves C, Spector TD, et al. Predicting brain age with deep learning from raw imaging data results in a reliable and heritable biomarker. NeuroImage. 2017;163:115–24.
Buchlak QD, Esmaili N, Leveque J-C, Farrokhi F, Bennett C, Piccardi M, et al. Machine learning applications to clinical decision support in neurosurgery: an artificial intelligence augmented systematic review. Neurosurgical review. 2019 Aug 17:1–9.
Tosetti P, Hicks RR, Theriault E, Phillips A, Koroshetz W, Draghia-Akli R, et al. Toward an international initiative for traumatic brain injury research. J Neurotrauma. 2013;30(14):1211–22.
Angus DC. Fusing Randomized Trials With Big Data: The Key to Self-learning Health Care Systems? JAMA. 2015;314(8):767.
Jabbarli R, Pierscianek D, Darkwah Oppong M, Sato T, Dammann P, Wrede KH, et al. Laboratory biomarkers of delayed cerebral ischemia after subarachnoid hemorrhage: a systematic review. Neurosurgical review. 2018 Oct 10:1–9.
Claassen J, Bernardini GL, Kreiter K, Bates J, Du YE, Copeland D, et al. Effect of cisternal and ventricular blood on risk of delayed cerebral ischemia after subarachnoid hemorrhage: the Fisher scale revisited. Stroke. 2001;32(9):2012–20.
Woitzik J, Dreier JP, Hecht N, Fiss I, Sandow N, Major S, et al. Delayed Cerebral Ischemia and Spreading Depolarization in Absence of Angiographic Vasospasm after Subarachnoid Hemorrhage. Journal of Cerebral Blood Flow & Metabolism. 2012;32(2):203–12.
Beqiri E, Smielewski P, Robba C, Czosnyka M, Cabeleira MT, Tas J, et al. Feasibility of individualised severe traumatic brain injury management using an automated assessment of optimal cerebral perfusion pressure: the COGiTATE phase II study protocol. BMJ Open. 2019;9(9):e030727.
Olsen L, Aisner D, McGinnis JM. Institute of Medicine (US). Roundtable on Evidence-Based Medicine. The Learning Healthcare System: Workshop Summary. Natl Academy Pr; 2007 Jun 1.
Marsolo K, Margolis PA, Forrest CB, Colletti RB, Hutton JJ. A Digital Architecture for a Network-Based Learning Health System: Integrating Chronic Care Management, Quality Improvement, and Research. eGEMs (Generating Evidence & Methods to improve patient outcomes). 2015;3(1):16.
Yu J, Kahn JM. Creating a Learning Healthcare System in the ICU. In: Vincent J-L, ed. Annual Update in Intensive Care and Emergency Medicine 2015. Springer, Cham. 2015;(2015):587–96.
Orphanidou C. A review of big data applications of physiological signal data. Biophys Rev. 2019;11(1):83–7.
HIPAA Journal. 47GB of Medical Records and Test Results Found in Unsecured Amazon S3 Bucket [Internet]. HIPAA Journal. 2017 Oct 11. Available from: https://www.hipaajournal.com/47gb-medical-records-unsecured-amazon-s3-bucket/. Accessed 10 Jan 2020.
Ienca M, Ferretti A, Hurst S, Puhan M, Lovis C, Vayena E. Considerations for ethics review of big data health research: A scoping review. Biemba G, editor. PLOS ONE. 2018;13(10):e0204937.
Copeland R. Google’s ‘Project Nightingale’ Gathers Personal Health Data on Millions of Americans. Wall Street Journal. 2019 Nov 11. Available from: https://www.wsj.com/articles/google-s-secret-project-nightingale-gathers-personal-health-data-on-millions-of-americans-11573496790. Accessed 9 Jan 2020.
Gaspard N, Hirsch LJ. Pitfalls in ictal EEG interpretation: Critical care and intracranial recordings. Neurology. 2013;80(Issue 1, Supplement 1):S26–42.
Edinburgh T, Smielewski P, Czosnyka M, Eglen SJ, Ercole A. DeepClean--self-supervised artefact rejection for intensive care waveform data using generative deep learning. arXiv. 2019 Aug; preprint arXiv:1908.03129. Accessed 30 Oct 2019.
Obermeyer Z, Emanuel EJ. Predicting the Future—Big Data, Machine Learning, and Clinical Medicine. N Engl J Med. 2016;375(13):1216–9.
Cabeleira M, Ercole A, Smielewski P. HDF5-Based Data Format for Archiving Complex Neuro-monitoring Data in Traumatic Brain Injury Patients. In: Heldt T, ed. Intracranial Pressure & Neuromonitoring XVI. Springer International Publishing; 2018 Mar; 121–5. Available from: http://link.springer.com/10.1007/978-3-319-65798-1_26. Accessed 29 Oct 2019.
Li X, Morgan PS, Ashburner J, Smith J, Rorden C. The first step for neuroimaging data analysis: DICOM to NIfTI conversion. Journal of Neuroscience Methods. 2016;264:47–56.
Maas AIR, Harrison-Felix CL, Menon D, Adelson PD, Balkin T, Bullock R, et al. Standardizing Data Collection in Traumatic Brain Injury. Journal of Neurotrauma. 2011;28(2):177–87.
Melinosky C, Yang S, Hu P, Li H, Miller CHT, Khan I, et al. Continuous Vital Sign Analysis to Predict Secondary Neurological Decline After Traumatic Brain Injury. Frontiers in Neurology. 2018 Sep 25;9:761.
Resnick B. Yes, artificial intelligence can be racist. Vox. 2019 Jan 24. Available from: https://www.vox.com/science-and-health/2019/1/23/18194717/alexandria-ocasio-cortez-ai-bias. Accessed 30 Oct 2019.
Caliskan A, Bryson JJ, Narayanan A. Semantics derived automatically from language corpora contain human-like biases. Science. 2017;356(6334):183–6.
Elmer J, Torres C, Aufderheide TP, Austin MA, Callaway CW, Golan E, et al. Association of early withdrawal of life-sustaining therapy for perceived neurological prognosis with mortality after cardiac arrest. Resuscitation. 2016;102:127–35.
Garton ALA, Gupta VP, Sudesh S, Zhou H, Christophe BR, Connolly ES. The Intracerebral Hemorrhage Score: Changing Perspectives on Mortality and Disability. World Neurosurgery. 2019 Dec 21. In Press.
Teich DA. Management AI: A Senate Bill Introduced To Begin Regulation Of Machine Learning Algorithms. Forbes. 2019 Apr 25. Available from: https://www.forbes.com/sites/davidteich/2019/04/25/management-ai-a-senate-bill-introduced-to-begin-regulation-of-machine-learning-algorithms/#3823f07079e6. Accessed 29 Oct 2019.
Ma H, Campbell BCV, Parsons MW, Churilov L, Levi CR, Hsu C, et al. Thrombolysis Guided by Perfusion Imaging up to 9 Hours after Onset of Stroke. New England Journal of Medicine. 2019;380(19):1795–803.
Albers GW, Marks MP, Kemp S, Christensen S, Tsai JP, Ortega-Gutierrez S, et al. Thrombectomy for Stroke at 6 to 16 Hours with Selection by Perfusion Imaging. New England Journal of Medicine. 2018;378(8):708–18.
Sahoo SS, Wei A, Valdez J, Wang L, Zonjy B, Tatsuoka C, et al. NeuroPigPen: A Scalable Toolkit for Processing Electrophysiological Signal Data in Neuroscience Applications Using Apache Pig. Frontiers in Neuroinformatics [Internet]. 2016 ;10. [cited 2019 Sep 6]. Available from: http://journal.frontiersin.org/Article/10.3389/fninf.2016.00018/abstract
Dias C, Silva MJ, Pereira E, Monteiro E, Maia I, Barbosa S, et al. Optimal Cerebral Perfusion Pressure Management at Bedside: A Single-Center Pilot Study. Neurocritical Care. 2015;23(1):92–102.
Woodfield R, Grant I, UK Biobank Stroke Outcomes Group, UK Biobank Follow-Up and Outcomes Working Group, Sudlow CLM. Accuracy of Electronic Health Record Data for Identifying Stroke Cases in Large-Scale Epidemiological Studies: A Systematic Review from the UK Biobank Stroke Outcomes Group. PLoS ONE. 2015;10(10):e0140533.
Chang TE, Lichtman JH, Goldstein LB, George MG. Accuracy of ICD-9-CM Codes by Hospital Characteristics and Stroke Severity: Paul Coverdell National Acute Stroke Program. J Am Heart Assoc. 2016 May 31;5(6):e003056.
Marik PE, Baram M, Vahid B. Does Central Venous Pressure Predict Fluid Responsiveness?*: A Systematic Review of the Literature and the Tale of Seven Mares. Chest. 2008;134(1):172–8.
Schmidt JM, Sow D, Crimmins M, Albers D, Agarwal S, Claassen J, et al. Heart Rate Variability for Preclinical Detection of Secondary Complications After Subarachnoid Hemorrhage. Neurocritical Care. 2014;20(3):382–9.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Foreman, B. Neurocritical Care: Bench to Bedside (Eds. Claude Hemphill, Michael James) Integrating and Using Big Data in Neurocritical Care. Neurotherapeutics 17, 593–605 (2020). https://doi.org/10.1007/s13311-020-00846-1
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13311-020-00846-1