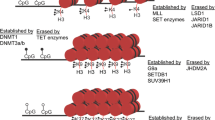
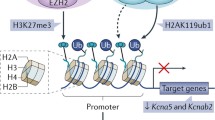
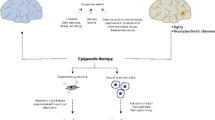
Abstract
This review highlights recent discoveries that have shaped the emerging viewpoints in the field of epigenetic influences in the central nervous system (CNS), focusing on the following questions: i) How is the CNS shaped during development when precursor cells transition into morphologically and molecularly distinct cell types, and is this event driven by epigenetic alterations?; ii) How do epigenetic pathways control CNS function?; iii) What happens to “epigenetic memory” during aging processes, and do these alterations cause CNS dysfunction?; iv) Can one restore normal CNS function by manipulating the epigenome using pharmacologic agents, and will this ameliorate aging-related neurodegeneration? These and other still unanswered questions remain critical to understanding the impact of multifaceted epigenetic machinery on the age-related dysfunction of CNS.




Similar content being viewed by others
References
Waddington CH. The strategy of the genes; a discussion of some aspects of theoretical biology. Allen & Unwin, London, 1957.
Waterland RA, Jirtle RL. Transposable elements: Targets for early nutritional effects on epigenetic gene regulation. Mol Cell Biol 2003;23:5293–5300.
Weaver IC, Cervoni N, Champagne FA, et al. Epigenetic programming by maternal behavior. Nat Neurosci 2004;7:847–854.
Qureshi IA, Mehler MF. Long non-coding RNAs: Novel targets for nervous system disease diagnosis and therapy. Neurotherapeutics 2013.
Tang B, Chang WL, Lanigan CM, Dean B, Sutcliffe JG, Thomas EA. Normal human aging and early-stage schizophrenia share common molecular profiles. Aging Cell 2009;8:339–342.
Narayan P, Dragunow M: Pharmacology of epigenetics in brain disorders. British journal of pharmacology (2010) 159(2):285–303.
Tollervey JR, Lunyak VV. Epigenetics: Judge, jury and executioner of stem cell fate. Epigenetics 2012;7:823–840.
Delatte B, Fuks F. Tet proteins: On the frenetic hunt for new cytosine modifications. Brief Funct Genomics 2013;12:191–204.
Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 1992;69:915–926.
Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases DNMT3a and DNMT3b are essential for de novo methylation and mammalian development. Cell 1999;99:247–257.
Wu H, Coskun V, Tao J, et al. DNMT3a-dependent nonpromoter DNA methylation facilitates transcription of neurogenic genes. Science 2010;329:444–448.
Feng J, Zhou Y, Campbell SL, et al. Dnmt1 and dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat Neurosci 2010;13:423–430.
Miller CA, Gavin CF, White JA, et al. Cortical DNA methylation maintains remote memory. Nat Neurosci 2010;13:664–666.
Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in x-linked mecp2, encoding methyl-CPG-binding protein 2. Nat Genet 1999;23:185–188.
Lunyak VV, Burgess R, Prefontaine GG, et al. Corepressor-dependent silencing of chromosomal regions encoding neuronal genes. Science 2002;298:1747–1752.
Szulwach KE, Li X, Li Y, et al. 5-hmc-mediated epigenetic dynamics during postnatal neurodevelopment and aging. Nat Neurosci 2011;14:1607–1616.
Wang T, Pan Q, Lin L, et al. Genome-wide DNA hydroxymethylation changes are associated with neurodevelopmental genes in the developing human cerebellum. Hum Mol Genet 2012;21:5500–5510.
Numata S, Ye T, Hyde TM, et al. DNA methylation signatures in development and aging of the human prefrontal cortex. Am J Hum Genet 2012;90:260–272.
Stadler F, Kolb G, Rubusch L, et al. Histone methylation at gene promoters is associated with developmental regulation and region-specific expression of ionotropic and metabotropic glutamate receptors in human brain. J Neurochem 2005;94:324–336.
Cheung I, Shulha HP, Jiang Y, et al. Developmental regulation and individual differences of neuronal h3k4me3 epigenomes in the prefrontal cortex. Proc Natl Acad Sci U S A 2010;107:8824–8829.
Wang CM, Tsai SN, Yew TW, Kwan YW, Ngai SM. Identification of histone methylation multiplicities patterns in the brain of senescence-accelerated prone mouse 8. Biogerontology 2010;11:87–102.
Lu T, Pan Y, Kao SY, et al. Gene regulation and DNA damage in the ageing human brain. Nature 2004;429:883–891.
Jakovcevski M, Akbarian S. Epigenetic mechanisms in neurological disease. Nat Med 2012;18:1194–1204.
Peleg S, Sananbenesi F, Zovoilis A, et al. Altered histone acetylation is associated with age-dependent memory impairment in mice. Science 2010;328:753–756.
Fischer A, Sananbenesi F, Mungenast A, Tsai LH. Targeting the correct hdac(s) to treat cognitive disorders. Trends Pharmacol Sci 2010;31:605–617.
Erraji-Benchekroun L, Underwood MD, Arango V, et al. Molecular aging in human prefrontal cortex is selective and continuous throughout adult life. Biol Psychiatry 2005;57:549–558.
Copray S, Huynh JL, Sher F, Casaccia-Bonnefil P, Boddeke E. Epigenetic mechanisms facilitating oligodendrocyte development, maturation, and aging. Glia 2009;57:1579–1587.
Ginsberg SD. Expression profile analysis of brain aging. In: Riddle DR (ed.) Brain aging: Models, methods, and mechanisms. CRS press. Boca Raton, FL, 2007, chaper 7, pp: 159–189.
Patterton D, Wolffe AP. Developmental roles for chromatin and chromosomal structure. Dev Biol 1996;173:2–13.
Bonisch C, Hake SB. Histone H2A variants in nucleosomes and chromatin: More or less stable? Nucleic Acids Res 2012;40:10719–10741.
Michod D, Bartesaghi S, Khelifi A, et al. Calcium-dependent dephosphorylation of the histone chaperone daxx regulates h3.3 loading and transcription upon neuronal activation. Neuron 2012;74:122–135.
Fernando RN, Eleuteri B, Abdelhady S, Nussenzweig A, Andang M, Ernfors P. Cell cycle restriction by histone h2ax limits proliferation of adult neural stem cells. Proc Natl Acad Sci U S A 2011;108:5837–5842.
Scaturro M, Nastasi T, Raimondi L, Bellafiore M, Cestelli A, Di Liegro I. H1(0) rna-binding proteins specifically expressed in the rat brain. J Biol Chem 1998;273:22788–22791.
Lindner H, Sarg B, Hoertnagl B, Helliger W. The microheterogeneity of the mammalian h1(0) histone. Evidence for an age-dependent deamidation. J Biol Chem 1998;273:13324–13330
Brown DT, Alexander BT, Sittman DB. Differential effect of h1 variant overexpression on cell cycle progression and gene expression. Nucleic Acids Res 1996;24:486–493.
Bantignies F, Cavalli G. Cellular memory and dynamic regulation of polycomb group proteins. Curr Opin Cell Biol 2006;18:275–283.
Heard E, Rougeulle C, Arnaud D, Avner P, Allis CD, Spector DL. Methylation of histone H3 at lys-9 is an early mark on the x chromosome during X inactivation. Cell 2001;107:727–738.
McKinnon PJ. Ataxia telangiectasia: New neurons and ATM. Trends Mol Med 2001;7:233–234.
Hon GC, Rajagopal N, Shen Y, et al. Epigenetic memory at embryonic enhancers identified in DNA methylation maps from adult mouse tissues. Nat Genet 2013; 45:1198–1206.
Santoro SW, Dulac C. The activity-dependent histone variant h2be modulates the life span of olfactory neurons. eLife 2012;1:e00070.
Rosenfeld MG, Lunyak VV, Glass CK. Sensors and signals: A coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Develop 2006;20:1405–1428.
Edelman LB, Fraser P. Transcription factories: Genetic programming in three dimensions. Curr Opin Genet Dev 2012;22:110–114.
Lanctot C, Cheutin T, Cremer M, Cavalli G, Cremer T. Dynamic genome architecture in the nuclear space: Regulation of gene expression in three dimensions. Nat Rev Genet 2007;8:104–115.
Sutherland H, Bickmore WA. Transcription factories: Gene expression in unions? Nat Rev Genet 2009;10:457–466.
Xu M, Cook PR. The role of specialized transcription factories in chromosome pairing. Biochim Biophys Acta 2008;1783:2155–2160.
Pina B and Suau P. Core histone variants and ubiquitinated histones 2a and 2b of rat cerebral cortex neurons. Biochem Biophys Res Commun 1985;133:505–510.
Duce JA, Smith DP, Blake RE, et al. Linker histone h1 binds to disease associated amyloid-like fibrils. J Mol Biol 2006;361:493–505.
Thakar A, Gupta P, Ishibashi T, et al. H2a.Z and h3.3 histone variants affect nucleosome structure: Biochemical and biophysical studies. Biochemistry 2009;48:10852–10857.
Jin C, Felsenfeld G. Nucleosome stability mediated by histone variants h3.3 and h2a.Z. Genes Develop 2007;21:1519–1529.
Crepaldi L, Policarpi C, Coatti A, et al. Binding of tfiiic to sine elements controls the relocation of activity-dependent neuronal genes to transcription factories. PLoS Genet 2013;9:e1003699.
Lunyak VV, Atallah M. Genomic relationship between sine retrotransposons, pol iii-pol ii transcription, and chromatin organization: The journey from junk to jewel. Biochem Cell Biol 2011; 89:495–504.
Lunyak VV. Boundaries. Boundaries…Boundaries??? Curr Opin Cell Biol 2008;20:281–287.
Wang J, Geesman GJ, Hostikka SL, et al. Inhibition of activated pericentromeric sine/alu repeat transcription in senescent human adult stem cells reinstates self-renewal. Cell Cycle 2011;10:3016–3030.
De Cecco M, Criscione SW, Peckham EJ, et al. Genomes of replicatively senescent cells undergo global epigenetic changes leading to gene silencing and activation of transposable elements. Aging Cell 2013;12:247–256.
Lunyak VV, Prefontaine GG, Nunez E, et al: Developmentally regulated activation of a sine b2 repeat as a domain boundary in organogenesis. Science 2007;317:248–251.
Hu Q, Kwon YS, Nunez E, et al. Enhancing nuclear receptor-induced transcription requires nuclear motor and lsd1-dependent gene networking in interchromatin granules. Proc Natl Acad Sci U S A 2008;105:19199–19204.
Surani MA. Reprogramming of genome function through epigenetic inheritance. Nature 2001;414:122–128.
Meshorer E, Yellajoshula D, George E, Scambler PJ, Brown DT, Misteli T. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev Cell 2006;10:105–116.
Solovei I, Grandi N, Knoth R, Folk B, Cremer T. Positional changes of pericentromeric heterochromatin and nucleoli in postmitotic Purkinje cells during murine cerebellum development. Cytogenet Genome Res 2004;105:302–310.
Bosch A, Suau P. Changes in core histone variant composition in differentiating neurons: The roles of differential turnover and synthesis rates. Eur J Cell Biol 1995;68:220–225.
Fontebasso AM, Liu XY, Sturm D, Jabado N. Chromatin remodeling defects in pediatric and young adult glioblastoma: A tale of a variant histone 3 tail. Brain Pathol 2013;23:210–216.
Wan LB, Bartolomei MS. Regulation of imprinting in clusters: Noncoding rnas versus insulators. Adv Genet 2008;61:207–223.
Tyler JK. Chromatin assembly. Cooperation between histone chaperones and atp-dependent nucleosome remodeling machines. Eur J Biochem 2002;269:2268–2274.
Ransom M, Dennehey BK, Tyler JK. Chaperoning histones during DNA replication and repair. Cell 2010;140:183–195.
Lopez MF, Tollervey J, Krastins B, et al. Depletion of nuclear histone h2a variants is associated with chronic DNA damage signaling upon drug-evoked senescence of human somatic cells. Aging 2012;4:823–842.
O'Sullivan RJ, Kubicek S, Schreiber SL, Karlseder J. Reduced histone biosynthesis and chromatin changes arising from a damage signal at telomeres. Nat Struct Mol Biol 2010;17:1218–1225.
Das C, Tyler JK, Churchill ME. The histone shuffle: Histone chaperones in an energetic dance. Trends Biochem Sci 2010;35:476–489.
Itoh T, Ausio J, Katagiri C. Histone h1 variants as sperm-specific nuclear proteins of rana catesbeiana, and their role in maintaining a unique condensed state of sperm chromatin. Mol Reprod Dev 1997;47:181–190.
Yang X, Khosravi-Far R, Chang HY, Baltimore D. Daxx, a novel fas-binding protein that activates jnk and apoptosis. Cell 1997;89:1067–1076.
Drane P, Ouararhni K, Depaux A, Shuaib M, Hamiche A. The death-associated protein daxx is a novel histone chaperone involved in the replication-independent deposition of h3.3. Genes Develop 2010;24:1253–1265.
Lewis PW, Elsaesser SJ, Noh KM, Stadler SC, Allis CD. DAXX is an h3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proc Natl Acad Sci U S A 2010;107:14075–14080.
Hollenbach AD, McPherson CJ, Mientjes EJ, Iyengar R, Grosveld G. DAXX and histone deacetylase ii associate with chromatin through an interaction with core histones and the chromatin-associated protein DEK. J Cell Sci 2002;115:3319–3330.
Kuo HY, Chang CC, Jeng JC, et al. Sumo modification negatively modulates the transcriptional activity of CREB-binding protein via the recruitment of DAXX. Proc Natl Acad Sci U S A 2005;10247:16973–16978.
Puto LA, Reed JC. Daxx represses relb target promoters via DNA methyltransferase recruitment and DNA hypermethylation. Genes Develop 2008;22:998–1010.
Salomoni P, Khelifi AF. DAXX: Death or survival protein? Trends Cell Biol 2006;16:97–104.
Goldberg AD, Banaszynski LA, Noh KM, et al. Distinct factors control histone variant h3.3 localization at specific genomic regions. Cell 2010;140:678–691.
Elsaesser SJ, Allis CD. HIRA and DAXX constitute two independent histone h3.3-containing predeposition complexes. Cold Spring Harb Symp Quant Biol 2010;75:27–34.
Schwartzentruber J, Korshunov A, Liu XY, et al. Driver mutations in histone h3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 2012;482:226–231.
Kamakaka RT, Biggins S. Histone variants: Deviants? Genes Develop 2005;19:295–310.
Old RW, Woodland HR. Histone genes: Not so simple after all. Cell 1984;38:624–626.
Henikoff S. Nucleosome destabilization in the epigenetic regulation of gene expression. Nat Rev Genet 2008;9:15–26.
Jin C, Zang C, Wei G, et al. H3.3/h2a.Z double variant-containing nucleosomes mark 'nucleosome-free regions' of active promoters and other regulatory regions. Nat Genet 2009;41:941–945.
Gessi M, Gielen GH, Hammes J, et al. H3.3 g34r mutations in pediatric primitive neuroectodermal tumors of central nervous system (CNS-PNET) and pediatric glioblastomas: Possible diagnostic and therapeutic implications? J Neurooncol 2013;112:67–72.
Lal A, Pan Y, Navarro F, et al. Mir-24-mediated downregulation of h2ax suppresses DNA repair in terminally differentiated blood cells. Nat Struct Mol Biol 2009;16:492–498.
Bassett A, Cooper S, Wu C, Travers A. The folding and unfolding of eukaryotic chromatin. Curr Opin Genet Dev 2009;19:159–165.
Kouzarides T. Chromatin modifications and their function. Cell 2007;128:693–705.
Strahl BD, Allis CD. The language of covalent histone modifications. Nature 2000;403:41–45.
Lee JS, Smith E, Shilatifard A. The language of histone crosstalk. Cell 2010;142:682-685.
Cheung P, Tanner KG, Cheung WL, Sassone-Corsi P, Denu JM, Allis CD. Synergistic coupling of histone h3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol Cell 2000;5:905–915.
Sassone-Corsi P, Mizzen CA, Cheung P, et al. Requirement of rsk-2 for epidermal growth factor-activated phosphorylation of histone h3. Science 1999;285:886–891.
Thomson S, Clayton AL, Hazzalin CA, Rose S, Barratt MJ, Mahadevan LC. The nucleosomal response associated with immediate-early gene induction is mediated via alternative map kinase cascades: Msk1 as a potential histone h3/hmg-14 kinase. EMBO J 1999;18:4779–4793.
Mahadevan LC, Willis AC, Barratt MJ. Rapid histone h3 phosphorylation in response to growth factors, phorbol esters, okadaic acid, and protein synthesis inhibitors. Cell 1991;65:775–783.
Chadee DN, Hendzel MJ, Tylipski CP, et al. Increased ser-10 phosphorylation of histone h3 in mitogen-stimulated and oncogene-transformed mouse fibroblasts. J Biol Chem 1999;274:24914–24920.
Nowak SJ, Corces VG. Phosphorylation of histone h3 correlates with transcriptionally active loci. Genes Develop 2000;14:3003–3013.
Crosio C, Fimia GM, Loury R, et al. Mitotic phosphorylation of histone h3: Spatio-temporal regulation by mammalian aurora kinases. Mol Cell Biol 2002;22:874–885.
Crosio C, Cermakian N, Allis CD, Sassone-Corsi P. Light induces chromatin modification in cells of the mammalian circadian clock. Nat Neurosci 2000;3:1241–1247.
Obrietan K, Impey S, Storm DR. Light and circadian rhythmicity regulate map kinase activation in the suprachiasmatic nuclei. Nat Neurosci 1998;1:693–700.
Travnickova-Bendova Z, Cermakian N, Reppert SM, Sassone-Corsi P. Bimodal regulation of mperiod promoters by creb-dependent signaling and clock/bmal1 activity. Proc Natl Acad Sci U S A 2002;99:7728–7733.
Goelet P, Castellucci VF, Schacher S, Kandel ER: The long and the short of long-term memory—a molecular framework. Nature 1986;322:419–422.
Wisden W, Errington ML, Williams S, et al. Differential expression of immediate early genes in the hippocampus and spinal cord. Neuron 1990;4:603–614.
Selcher JC, Nekrasova T, Paylor R, Landreth GE, Sweatt JD. Mice lacking the erk1 isoform of map kinase are unimpaired in emotional learning. Learn Mem 2001;8:11–19.
Rosenblum K, Futter M, Jones M, Hulme EC, Bliss TV. Erki/ii regulation by the muscarinic acetylcholine receptors in neurons. J Neurosci 2000;20:977–985.
Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature 2007;447:433–440.
Nestler EJ. Epigenetic mechanisms of drug addiction. Neuropharmacology 2013.
Camelo S, Iglesias AH, Hwang D, et al. Transcriptional therapy with the histone deacetylase inhibitor trichostatin a ameliorates experimental autoimmune encephalomyelitis. J Neuroimmunol 2005;164:10–21.
Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature 2007;447:425–432.
Pirooznia SK, Elefant F. Targeting specific hats for neurodegenerative disease treatment: Translating basic biology to therapeutic possibilities. Front Cell Neurosci 2013;7:30.
Cao X, Sudhof TC. A transcriptionally [correction of transcriptively] active complex of app with fe65 and histone acetyltransferase tip60. Science 2001;293:115–120.
Minopoli G, Gargiulo A, Parisi S, Russo T. Fe65 matters: New light on an old molecule. IUBMB Life 2012;64:936–942.
Adamec E, Vonsattel JP, Nixon RA. DNA strand breaks in Alzheimer's disease. Brain Res 1999;849:67–77.
Kim D, Frank CL, Dobbin MM, et al. Deregulation of HDAC1 by p25/CDK5 in neurotoxicity. Neuron 2008;60:803–817.
Marambaud P, Wen PH, Dutt A, et al. A cbp binding transcriptional repressor produced by the PS1/epsilon-cleavage of n-cadherin is inhibited by PS1 FAD mutations. Cell 2003;114:635–645.
Kim D, Nguyen MD, Dobbin MM, et al. Sirt1 deacetylase protects against neurodegeneration in models for alzheimer's disease and amyotrophic lateral sclerosis. EMBO J 2007;26:3169–3179.
Donmez G, Wang D, Cohen DE, Guarente L. Sirt1 suppresses beta-amyloid production by activating the alpha-secretase gene adam10. Cell 2010;142:320–332.
Rouaux C, Jokic N, Mbebi C, Boutillier S, Loeffler JP, Boutillier AL. Critical loss of CBP/p300 histone acetylase activity by caspase-6 during neurodegeneration. EMBO J 2003;22:6537–6549.
Ricobaraza A, Cuadrado-Tejedor M, Perez-Mediavilla A, Frechilla D, Del Rio J, Garcia-Osta A. Phenylbutyrate ameliorates cognitive deficit and reduces tau pathology in an alzheimer's disease mouse model. Neuropsychopharmacology 2009;34:1721–1732.
Kontopoulos E, Parvin JD, Feany MB. Alpha-synuclein acts in the nucleus to inhibit histone acetylation and promote neurotoxicity. Hum Mol Genet 2006;15:3012–3023.
Outeiro TF, Kontopoulos E, Altmann SM, et al. Sirtuin 2 inhibitors rescue alpha-synuclein-mediated toxicity in models of parkinson's disease. Science 2007;317:516–519.
Chen PS, Wang CC, Bortner CD, et al. Valproic acid and other histone deacetylase inhibitors induce microglial apoptosis and attenuate lipopolysaccharide-induced dopaminergic neurotoxicity. Neuroscience 2007;149:203–212.
Wang Y, Wang X, Liu L, Wang X. HDAC inhibitor trichostatin A-inhibited survival of dopaminergic neuronal cells. Neurosci Lett 2009;467:212–216.
Zee BM, Levin RS, Xu B, LeRoy G, Wingreen NS, Garcia BA. In vivo residue-specific histone methylation dynamics. J Biol Chem 2010;285:3341–3350.
Barth TK, Imhof A. Fast signals and slow marks: The dynamics of histone modifications. Trends Biochem Sci 2010;35:618–626.
Peter CJ, Akbarian S. Balancing histone methylation activities in psychiatric disorders. Trends Mol Med 2011;17:372–379.
Crosio C, Heitz E, Allis CD, Borrelli E, Sassone-Corsi P. Chromatin remodeling and neuronal response: Multiple signaling pathways induce specific histone h3 modifications and early gene expression in hippocampal neurons. J Cell Sci 2003;116:4905–4914.
Hunter RG, Murakami G, Dewell S, et al. Acute stress and hippocampal histone H3 lysine 9 trimethylation, a retrotransposon silencing response. Proc Natl Acad Sci U S A 2012;109:17657–17662.
Hunter RG, McCarthy KJ, Milne TA, Pfaff DW, McEwen BS. Regulation of hippocampal h3 histone methylation by acute and chronic stress. Proc Natl Acad Sci U S A 2009;106:20912–20917.
Birney E, Stamatoyannopoulos JA, Dutta A, et al. Identification and analysis of functional elements in 1% of the human genome by the encode pilot project. Nature 2007;447:799–816.
Kazazian HH, Jr, Goodier JL. Line drive. Retrotransposition and genome instability. Cell 2002;110:277–280.
Perron H, Lang A. The human endogenous retrovirus link between genes and environment in multiple sclerosis and in multifactorial diseases associating neuroinflammation. Clin Rev Allergy Immunol 2010;39:51–61.
Ponicsan SL, Kugel JF, Goodrich JA. Genomic gems: Sine RNAs regulate mRNA production. Curr Opin Genet Dev 2010;20:149–155.
Muotri AR, Marchetto MC, Coufal NG, et al. L1 retrotransposition in neurons is modulated by mecp2. Nature 2010;468:443–446.
Ryu H, Lee J, Hagerty SW, et al. Eset/setdb1 gene expression and histone h3 (k9) trimethylation in huntington's disease. Proc Natl Acad Sci U S A 2006;103:19176–19181.
Rea S, Eisenhaber F, O'Carroll D, et al. Regulation of chromatin structure by site-specific histone h3 methyltransferases. Nature 2000;406:593–599.
Hu Y, Chopra V, Chopra R, et al. Transcriptional modulator h2a histone family, member y (h2afy) marks huntington disease activity in man and mouse. Proc Natl Acad Sci U S A 2011;108:17141–17146.
Stack EC, Del Signore SJ, Luthi-Carter R, et al. Modulation of nucleosome dynamics in huntington's disease. Hum Mol Genet 2007;16:1164–1175.
Ayyanathan K, Lechner MS, Bell P, et al. Regulated recruitment of hp1 to a euchromatic gene induces mitotically heritable, epigenetic gene silencing: A mammalian cell culture model of gene variegation. Genes Develop 2003;17:1855–1869.
Jakobsson J, Cordero MI, Bisaz R, et al. Kap1-mediated epigenetic repression in the forebrain modulates behavioral vulnerability to stress. Neuron 2008;60:818–831.
Jiang Y, Jakovcevski M, Bharadwaj R, et al. SETDB1 histone methyltransferase regulates mood-related behaviors and expression of the nmda receptor subunit NR2B. J Neurosci 2010;30:7152–7167.
Penner MR, Roth TL, Chawla MK, et al. Age-related changes in arc transcription and DNA methylation within the hippocampus. Neurobiol Aging 2011;32:2198–2210.
Peters AH, Schubeler D. Methylation of histones: Playing memory with DNA. Curr Opin Cell Biol 2005;17:230–238.
Gupta-Agarwal S, Franklin AV, Deramus T, et al. G9a/glp histone lysine dimethyltransferase complex activity in the hippocampus and the entorhinal cortex is required for gene activation and silencing during memory consolidation. J Neurosci 2012;32:5440–5453.
Shinkai Y, Tachibana M: H3k9 methyltransferase G9a and the related molecule GLP. Genes Develop 2011;25:781–788.
Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol 2005;6:838–849.
Acknowledgments
YZ is a Buck Institute Research Fellow, and experiments from VVL laboratory are supported by NIH R21 AG043921 and Buck Institute Trust funds. We apologize to our colleagues for omission of so many important research contributions owing to the space constraints of this review. We thank J. Jenkins and L. Hankock for help with the manuscript preparation. We acknowledge members of the Lunyak laboratory for helpful discussions. No real or perceived conflict of interest is declared. Full conflict of interest disclosures are available in the electronic supplementary material for this article.
Required Author Forms Disclosure forms provided by the authors are available with the online version of this article.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 1224 kb)
Rights and permissions
About this article
Cite this article
Zhao, YQ., Jordan, I.K. & Lunyak, V.V. Epigenetics Components of Aging in the Central Nervous System. Neurotherapeutics 10, 647–663 (2013). https://doi.org/10.1007/s13311-013-0229-y
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13311-013-0229-y