Abstract
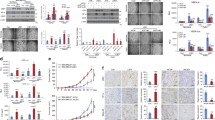
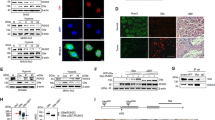
Hypoxia-inducible factor 2α (HIF2α) plays critical roles in cancer progression. Although the mechanisms of HIF2α translation and degradation have been well studied, the mechanism for HIF2α regulation at transcriptional level is still not fully understood. Here, we present evidence that DNA methylation in promoter contributes to transcription of EPAS1 coding HIF2α. Methylated CpG binding protein 3 (MBD3) contributes to the intricate regulatory mechanism. We showed that MBD3 bound to the EPAS1 promoter in breast cancer cells and amplified EPAS1 transcription through demethylating CpG located around transcriptional start site in MDA-MB-468 cells. This enabled MDA-MB-468 cells to activate HIF2α-mediated angiogenesis. However, in 7860 cells, the demethylation function of MBD3 on EPAS1 was not observed because of the poor methylated-CpG promoter. Nevertheless, depletion of MBD3 induced by shRNA decreased EPAS1 transcription and therefore decreased HIF2α-mediated cellular response in both MDA-MB-468 and 7860 cancer cells. These results indicated that the endogenous MBD3 was involved in regulating the transcription and therefore the transcriptional activities of HIF2α, suggesting that MBD3 may be a potential therapeutic target of tumor.






Similar content being viewed by others
References
Yoshida Y, Takahashi K, Okita K, Ichisaka T, Yamanaka S. Hypoxia enhances the generation of induced pluripotent stem cells. Cell Stem Cell. 2009;5(3):237–41. doi:10.1016/j.stem.2009.08.001.
Srinivasan S, Chitalia V, Meyer RD, Hartsough E, Mehta M, Harrold I, et al. Hypoxia-induced expression of phosducin-like 3 regulates expression of VEGFR-2 and promotes angiogenesis. Angiogenesis. 2015;18(4):449–62. doi:10.1007/s10456-015-9468-3.
De Miguel MP, Alcaina Y, de la Maza DS, Lopez-Iglesias P. Cell metabolism under microenvironmental low oxygen tension levels in stemness, proliferation and pluripotency. Curr Mol Med. 2015;15(4):343–59. doi:10.2174/1566524015666150505160406.
Vogler M, Vogel S, Krull S, Farhat K, Leisering P, Lutz S, et al. Hypoxia modulates fibroblastic architecture, adhesion and migration: a role for HIF-1alpha in cofilin regulation and cytoplasmic actin distribution. PLoS One. 2013;8(7):e69128. doi:10.1371/journal.pone.0069128.
Dudas J, Schartinger VH, Romani A, Schweigl G, Kordsmeyer K, Marta PI, et al. Cell cycle association and hypoxia regulation of excision repair cross complementation group 1 protein (ERCC1) in tumor cells of head and neck cancer. Tumour Biol. 2014;35(8):7807–19. doi:10.1007/s13277-014-2001-2.
van den Beucken T, Koch E, Chu K, Rupaimoole R, Prickaerts P, Adriaens M, et al. Hypoxia promotes stem cell phenotypes and poor prognosis through epigenetic regulation of DICER. Nat Commun. 2014;5:5203. doi:10.1038/ncomms6203.
Gordan JD, Bertout JA, Hu CJ, Diehl JA, Simon MC. HIF-2alpha promotes hypoxic cell proliferation by enhancing c-myc transcriptional activity. Cancer Cell. 2007;11(4):335–47. doi:10.1016/j.ccr.2007.02.006.
Bertout JA, Majmundar AJ, Gordan JD, Lam JC, Ditsworth D, Keith B, et al. HIF2alpha inhibition promotes p53 pathway activity, tumor cell death, and radiation responses. Proc Natl Acad Sci U S A. 2009;106(34):14391–6. doi:10.1073/pnas.0907357106.
Takeda N, O’Dea EL, Doedens A, Kim JW, Weidemann A, Stockmann C, et al. Differential activation and antagonistic function of HIF-{alpha} isoforms in macrophages are essential for NO homeostasis. Genes Dev. 2010;24(5):491–501. doi:10.1101/gad.1881410.
Kim WY, Perera S, Zhou B, Carretero J, Yeh JJ, Heathcote SA, et al. HIF2 alpha cooperates with RAS to promote lung tumorigenesis in mice. J Clin Investig. 2009;119(10):3182. doi:10.1172/Jci38443c1.
Devailly DVG, Augert A, Calvé BL, Ferrand M, Pigny P, Payen L, et al. Repression of PLA2R1 by c-MYC and HIF-2alpha promotes cancer growth. Oncotarget. 2014;5. doi:10.18632/oncotarget.1681.
Raspaglio G, Petrillo M, Martinelli E, Li Puma DD, Mariani M, De Donato M, et al. Sox9 and Hif-2alpha regulate TUBB3 gene expression and affect ovarian cancer aggressiveness. Gene. 2014;542(2):173–81. doi:10.1016/j.gene.2014.03.037.
Li Z, Bao S, Wu Q, Wang H, Eyler C, Sathornsumetee S, et al. Hypoxia-inducible factors regulate tumorigenic capacity of glioma stem cells. Cancer Cell. 2009;15(6):501–13. doi:10.1016/j.ccr.2009.03.018.
Heddleston JM, Li Z, McLendon RE, Hjelmeland AB, Rich JN. The hypoxic microenvironment maintains glioblastoma stem cells and promotes reprogramming towards a cancer stem cell phenotype. Cell Cycle. 2009;8(20):3274–84. doi:10.4161/cc.8.20.9701.
Micucci C, Matacchione G, Valli D, Orciari S, Catalano A. HIF2alpha is involved in the expansion of CXCR4-positive cancer stem-like cells in renal cell carcinoma. Br J Cancer. 2015;113(8):1178–85. doi:10.1038/bjc.2015.338.
Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69(6):915–26. doi:10.1016/0092-8674(92)90611-F.
Pastor WA, Aravind L, Rao A. TETonic shift: biological roles of TET proteins in DNA demethylation and transcription. Nat Rev Mol Cell Biol. 2013;14(6):341–56. doi:10.1038/nrm3589.
Shahrzad S, Bertrand K, Minhas K, Coomber BL. Induction of DNA hypomethylation by tumor hypoxia. Epigenetics. 2007;2(2):119–25. doi:10.4161/epi.2.2.4613.
Liu Q, Liu L, Zhao Y, Zhang J, Wang D, Chen J, et al. Hypoxia induces genomic DNA demethylation through the activation of HIF-1alpha and transcriptional upregulation of MAT2A in hepatoma cells. Mol Cancer Ther. 2011;10(6):1113–23. doi:10.1158/1535-7163.MCT-10-1010.
Pal A, Srivastava T, Sharma MK, Mehndiratta M, Das P, Sinha S, et al. Aberrant methylation and associated transcriptional mobilization of Alu elements contributes to genomic instability in hypoxia. J Cell Mol Med. 2010;14(11):2646–54. doi:10.1111/j.1582-4934.2009.00792.x.
Rawluszko-Wieczorek AA, Horbacka K, Krokowicz P, Misztal M, Jagodzinski PP. Prognostic potential of DNA methylation and transcript levels of HIF1A and EPAS1 in colorectal cancer. Mol Cancer Res. 2014;12(8):1112–27. doi:10.1158/1541-7786.MCR-14-0054.
Yoo S, Takikawa S, Geraghty P, Argmann C, Campbell J, Lin L, et al. Integrative analysis of DNA methylation and gene expression data identifies EPAS1 as a key regulator of COPD. PLoS Genet. 2015;11(1):e1004898. doi:10.1371/journal.pgen.1004898.
Yildirim O, Li R, Hung JH, Chen PB, Dong X, Ee LS, et al. Mbd3/NURD complex regulates expression of 5-hydroxymethylcytosine marked genes in embryonic stem cells. Cell. 2011;147(7):1498–510. doi:10.1016/j.cell.2011.11.054.
Spruijt CG, Gnerlich F, Smits AH, Pfaffeneder T, Jansen PW, Bauer C, et al. Dynamic readers for 5-(hydroxy)methylcytosine and its oxidized derivatives. Cell. 2013;152(5):1146–59. doi:10.1016/j.cell.2013.02.004.
Brown SE, Suderman MJ, Hallett M, Szyf M. DNA demethylation induced by the methyl-CpG-binding domain protein MBD3. Gene. 2008;420(2):99–106. doi:10.1016/j.gene.2008.05.009.
Gunther K, Rust M, Leers J, Boettger T, Scharfe M, Jarek M, et al. Differential roles for MBD2 and MBD3 at methylated CpG islands, active promoters and binding to exon sequences. Nucleic Acids Res. 2013;41(5):3010–21. doi:10.1093/nar/gkt035.
Cui Y, Cho IH, Chowdhury B, Irudayaraj J. Real-time dynamics of methyl-CpG-binding domain protein 3 and its role in DNA demethylation by fluorescence correlation spectroscopy. Epigenetics. 2013;8(10):1089–100. doi:10.4161/epi.25958.
Aranda E, Owen GI. A semi-quantitative assay to screen for angiogenic compounds and compounds with angiogenic potential using the EA.hy926 endothelial cell line. Biol Res. 2009;42(3):377–89. doi:/S0716-97602009000300012.
Lachance G, Uniacke J, Audas TE, Holterman CE, Franovic A, Payette J, et al. DNMT3a epigenetic program regulates the HIF-2alpha oxygen-sensing pathway and the cellular response to hypoxia. Proc Natl Acad Sci U S A. 2014;111(21):7783–8. doi:10.1073/pnas.1322909111.
Corbet C, Draoui N, Polet F, Pinto A, Drozak X, Riant O, et al. The SIRT1/HIF2alpha axis drives reductive glutamine metabolism under chronic acidosis and alters tumor response to therapy. Cancer Res. 2014;74(19):5507–19. doi:10.1158/0008-5472.CAN-14-0705.
Fatrai S, Wierenga ATJ, Daenen SMGJ, Vellenga E, Schuringa JJ. Identification of HIF2 alpha as an important STAT5 target gene in human hematopoietic stem cells. Blood. 2011;117(12):3320–30. doi:10.1182/blood-2010-08-303669.
Purdue MP, Johansson M, Zelenika D, Toro JR, Scelo G, Moore LE, et al. Genome-wide association study of renal cell carcinoma identifies two susceptibility loci on 2p21 and 11q13.3. Nat Genet. 2011;43(1):60–5. doi:10.1038/ng.723.
Putra AC, Eguchi H, Lee KL, Yamane Y, Gustine E, Isobe T, et al. The A allele at rs13419896 of EPAS1 is associated with enhanced expression and poor prognosis for non-small cell lung cancer. PLoS One. 2015;10(8):e0134496. doi:10.1371/journal.pone.0134496.
Joshi S, Singh AR, Zulcic M, Durden DL. A macrophage-dominant PI3K isoform controls hypoxia-induced HIF1alpha and HIF2alpha stability and tumor growth, angiogenesis, and metastasis. Mol Cancer Res. 2014;12(10):1520–31. doi:10.1158/1541-7786.MCR-13-0682.
Mohlin S, Hamidian A, von Stedingk K, Bridges E, Wigerup C, Bexell D, et al. PI3K-mTORC2 but not PI3K-mTORC1 regulates transcription of HIF2A/EPAS1 and vascularization in neuroblastoma. Cancer Res. 2015. doi:10.1158/0008-5472.CAN-15-0708.
Moniz S, Bandarra D, Biddlestone J, Campbell KJ, Komander D, Bremm A, et al. Cezanne regulates E2F1-dependent HIF2alpha expression. J Cell Sci. 2015;128(16):3082–93. doi:10.1242/jcs.168864.
Hendrich B, Guy J, Ramsahoye B, Wilson VA, Bird A. Closely related proteins MBD2 and MBD3 play distinctive but interacting roles in mouse development. Genes Dev. 2001;15(6):710–23. doi:10.1101/gad.194101.
Rais Y, Zviran A, Geula S, Gafni O, Chomsky E, Viukov S, et al. Deterministic direct reprogramming of somatic cells to pluripotency. Nature. 2013;502(7469):65–70. doi:10.1038/nature12587.
Cramer JM, Scarsdale JN, Walavalkar NM, Buchwald WA, Ginder GD, Williams Jr DC. Probing the dynamic distribution of bound states for methylcytosine-binding domains on DNA. J Biol Chem. 2014;289(3):1294–302. doi:10.1074/jbc.M113.512236.
Iurlaro M, Ficz G, Oxley D, Raiber EA, Bachman M, Booth MJ, et al. A screen for hydroxymethylcytosine and formylcytosine binding proteins suggests functions in transcription and chromatin regulation. Genome Biol. 2013;14(10):R119. doi:10.1186/gb-2013-14-10-r119.
Cui Y, Irudayaraj J. Dissecting the behavior and function of MBD3 in DNA methylation homeostasis by single-molecule spectroscopy and microscopy. Nucleic Acids Res. 2015;43(6):3046–55. doi:10.1093/nar/gkv098.
Brown SE, Szyf M. Epigenetic programming of the rRNA promoter by MBD3. Mol Cell Biol. 2007;27(13):4938–52. doi:10.1128/MCB.01880-06.
Sansom OJ, Maddison K, Clarke AR. Mechanisms of disease: methyl-binding domain proteins as potential therapeutic targets in cancer. Nat Clin Pract Oncol. 2007;4(5):305–15. doi:10.1038/ncponc0812.
Crook JM, Dunn NR, Colman A. Repressed by a NuRD. Nat Cell Biol. 2006;8(3):212–4. doi:10.1038/ncb0306-212.
Peng L, Li Y, Xi Y, Li W, Li J, Lv R, et al. Methyl-CpG-binding domain protein 3-like 2 (MBD3L2) promotes Tet2 enzymatic activity for mediating 5mC oxidation. J Cell Sc. 2016. doi:10.1242/jcs.179044.
Acknowledgments
This work was supported by grant from the National Natural Science Foundation of China (No. 81372319) to Luo Gu; the National Natural Science Foundation of China (No. 81201614), the Natural Science Foundation of Jiangsu province (No. BK2012839), the Postdoctoral Science Foundation of China (No. 2012 T50511), and the High-Level Talents in Six Industries of Jiangsu Province (No. JY-020) to Jun Du; a grant from the College Graduates Research and Innovation Program of Jiangsu Province (No. JX22013315) to Jie Cui; and a Project Funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
None
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOCX 13 kb)
Rights and permissions
About this article
Cite this article
Cui, J., Duan, B., Zhao, X. et al. MBD3 mediates epigenetic regulation on EPAS1 promoter in cancer. Tumor Biol. 37, 13455–13467 (2016). https://doi.org/10.1007/s13277-016-5237-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-016-5237-1