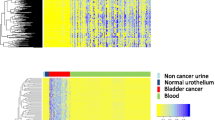
Abstract
Early screening for bladder cancer (BC) holds the key to combat and control the increasing global burden of BC mortality. We presented a simple approach to characterize, analyze, and validate a panel of biomarkers in BC and their relationship to bilharziasis. We investigated voided urine and blood samples from patients with bladder cancer (n = 94), benign bladder lesions (n = 60), and age-matched normal controls (n = 56). This study was divided into the following phases. (1) We analyzed the expression of urinary Hyaluronoglucosaminidase 1 (HYAL1) protein in BC and control samples by zymography. (2) We performed bioinformatics analysis to retrieve a set of epigenetic regulators of HYAL1. (3) This set of three selected genes [long non-coding RNA-urothelial cancer associated 1(lncRNA-UCA1), microRNA-210, and microRNA-96] was then analyzed in the same urine samples used in phase I by quantitative real-time PCR. (4) A high reproducibility of gene selection results was also determined from statistical validation. The urinary expression of HYAL1 protein and its epigenetic regulators were higher in BC patients (P < .001). The receiver-operating characteristic curve analyses demonstrated that each one had good sensitivity and specificity for distinguishing BC patients from non-BC ones (HYAL1, 89.4 and 91.2 %; miR-210, 76.6 and 93 %; miR-96, 76.6 and 89.4 %; and lncRNA-UCA1, 91.5 and 96.5 %). There was a significant positive correlation between HYAL1 and the selected epigenetic biomarkers. The performance of this urine biomarker panel reached 100 % sensitivity and 89.5 % specificity for bladder cancer diagnosis.


Similar content being viewed by others
Abbreviations
- BC:
-
Bladder cancer
- HYAL1:
-
Hyaluronoglucosaminidase 1
- kDa:
-
Kilodalton
- LncRNA:
-
Long non-coding RNA
- MiR:
-
Micro-RNA
- qPCR:
-
Quantitative polymerase chain reaction
- RT-PCR:
-
Reverse transcription polymerase chain reaction
- SCC:
-
Squamous cell carcinoma
- TCC:
-
Transitional cell carcinoma
- UCA1:
-
Urothelial cancer associated 1
References
Siegel R, Jiemin M, Zhaohui Z, Jemal A. Cancer statistics 2014. A Cancer Journal for Clinicians. 2014;64(1):15422–4863.
Sanchez C, Lachaize C, Janody F, Bellon B, Roder L, et al. Grasping at molecular interactions and genetic networks in Drosophila melanogaster using FlyNets, an Internet database. Nucleic Acids Res. 1999;27:89–94.
Lancashire LJ, Lemetre C, Ball GR. An introduction to artificial neural networks in bioinformatics—application to complex microarray and mass spectrometry datasets in cancer studies. Brief Bioinform. 2009;10:315–29.
Fraser JR, Laurent TC, Laurent UB. Hyaluronan: its nature, distribution, functions and turnover. J Intern Med. 1997;242:27–33.
Eissa S, Shehata H, Mansour A, Esmat M, El-Ahmady O. Detection of hyaluronidase RNA and activity in urine of schistosomal and non-schistosomal bladder cancer. Med Oncol. 2012;29:3345–51.
Miah S, Dudziec E, Drayton RM, et al. An evaluation of urinary microRNA reveals a high sensitivity for bladder cancer. Br J Cancer. 2012;107(1):123–8. doi:10.1038/bjc.2012.221.
Han Y, Liu Y, Nie L, et al. Inducing cell proliferation inhibition, apoptosis, and motility reduction by silencing long noncoding ribonucleic acid metastasis-associated lung adenocarcinoma transcript 1 in urothelial carcinoma of the bladder. Urology. 2013;81(209):e1–7.
Noon AP, Catto JW. Noncoding RNA in bladder cancer: a specific focus upon high-risk nonmuscle invasive disease. Curr Opin Urol. 2014;24(5):506–11.
Srivastava AK, Singh PK, Rath SK, Dalela D, Goel MM, Bhatt ML. Appraisal of diagnostic ability of UCA1 as a biomarker of carcinoma of the urinary bladder. Tumour Biol. 2014;15.
Qin Q, Furong W, Baosheng L. Multiple functions of hypoxia-regulated miR-210 in cancer. Cancer Res J Exp Clin. 2014;33:50. doi:10.1186/1756-9966-33-50.
Wang Y, Luo H, Li Y, Chen T, Wu S, Yang L. hsa-miR-96 up-regulates (MAP4K1 and IRS1 and may function as a promising diagnostic marker in human bladder urothelial carcinomas. Mol Med. 2012;5(1):260–5.
Jamshidian H, Hashemi M, Nowroozi MR, Ayati M, Bonyadi M, NajjaranTousi V. Sensitivity and specificity of urinary hyaluronic acid and hyaluronidase in detection of bladder transitional cell carcinoma. Urol J. 2014;11(1):1232–7.
Edge SB, Byrd DR, Compton CC, et al. AJCC cancer staging manual. 7th ed. New York: Springer; 2010. p. 497–505.
NCCN Clinical Practice Guidelines in Oncology: Bladder Cancer V. 2.2013. Available at http://bit.ly/jXgCvZ.Accessed September 5, 2013.
Eble JN, Sauter G, Epstein JI, Sesterhenn I, editors. World Health Organization classification of tumors pathology and genetics: tumors of the urinary system and male genital organs. Lyon: IARC Press; 2004.
Gui M, Idris MA, Shi YE, Muhling A, Ruppel A. Reactivity of Schistosoma japonicum and S. mansoni antigen preparations in indirect haemagglutination (IHA) with sera of patients with homologous and heterogonous schistosomiasis. Ann Trop Med Parasitol. 1991;85:599–604.
Papageorgakopoulou N, Vynios DH, Karayanni K, Maras A, Papapetropoulou M. Electrophoretic analysis of hydrolytic enzymes of Escherichia coli cells starved in seawater and drinking water: comparison of gelatinolytic, caseinolytic, phosphohydrolytic and hyaluronolytic activities. Microbiol Res. 1997;152:299–305.
Paraskevopoulou MD, Georgakilas G, Kostoulas Net al. DIANA-microT web server v5.0: service integration into miRNA functional analysis workflows. Nucleic Acids Res. 2013 Jul;41(Web Server issue):W169-73. Available at http://diana.cslab.ece.ntua.gr/pathways/
Thompson JD, Higgins DG, Gibson TJ. Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–80. CLUSTAL W.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−delta delta C (T)) method. Methods. 2001;25:402–8.
Nossier AI, Eissa S, Ismail MF, Hamdy MA, Azzazy HM. Direct detection of hyaluronidase in urine using cationic gold nanoparticles: a potential diagnostic test for bladder cancer. Biosens Bioelectron. 2014;54:7–14. doi:10.1016/j.bios.2013.10.024.
Place RF, Li LC, Pookot D, Noonan EJ, Dahiya R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc Natl Acad Sci U S A. 2008;105(5):1608–13. doi:10.1073/pnas.0707594105.
Zhu L, Liu J, Cheng G. Role of microRNAs in schistosomes and schistosomiasis. Front Cell Infect Microbiol. 2014;4:165. doi:10.3389/fcimb.2014.00165. eCollection 2014.
Acknowledgments
This work was supported by Ain Shams University Research Projects 2013–14. The authors are grateful to Dr. Nahla M. Awad, Ass. Prof. of Pathology at Early Cancer Detection Unit, Faculty of Medicine, Ain Shams University, for her help in the cytological examinations of all investigated urine samples. All authors have read the journal’s policy on disclosure of potential conflicts of interest. The authors have no conflict of interest. All authors have read the journal’s authorship agreement and that the manuscript has been reviewed by and approved by all named authors.
Conflicts of interest
None.
Authors’ contributions
Eissa S has participated in the design of the study, carried out data analysis, involved in drafting the manuscript or revising and has given final approval of the version to be published. Matboli M has performed bioinformatic analysis, practical work, participated in the design of the study, and performed the statistical analysis. Essawy N participated in the study design and involved in drafting the manuscript or revising. Youssef M. Kotb has provided us with urine, blood samples, and patient data and has given final approval of the version to be published. All authors read and approved the final manuscript.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary materials
Below is the link to the electronic supplementary material.
ESM 1
(DOC 1082 kb).
Rights and permissions
About this article
Cite this article
Eissa, S., Matboli, M., Essawy, N.O.E. et al. Integrative functional genetic-epigenetic approach for selecting genes as urine biomarkers for bladder cancer diagnosis. Tumor Biol. 36, 9545–9552 (2015). https://doi.org/10.1007/s13277-015-3722-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-015-3722-6