Abstract
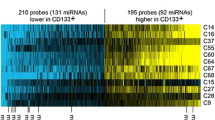
This study aimed to identify carcinogenic potential-related molecular mechanisms in cancer stem cells (CSCs) in lung cancer. CD133+ and CD133− subpopulations were sorted from A549 cells using magnetic-activated cell sorting. The abilities to form sphere and clone, proliferate, migrate, and invade were compared between CD133+ and CD133− cells, as well as drug sensitivity. Thereafter, microRNA (miRNA) profiles were performed to identify differentially expressed miRNAs between CD133+ and CD133− subpopulation. Following, bioinformatic methods were used to predict target genes for differentially expressed miRNAs and perform enrichment analysis. Furthermore, the mammalian target of rapamycin (mTOR) signaling pathways and CSC property-associated signaling pathways were explored and visualized in regulatory network among competitive endogenous RNA (ceRNA), miRNA, and target gene. CD133+ subpopulation showed greater oncogenic potential than CD133− subpopulation. In all, 14 differentially expressed miRNAs were obtained and enriched in 119 pathways, including five upregulated (hsa-miR-23b-3p, -23a-3p, -15b-5p, -24-3p, and -4734) and nine downregulated (hsa-miR-1246, -30b-5p, -5096, -6510-5p, has-miR-7110-5p, -7641, -3197, -7108-5p, and -6791-5p). For mTOR signaling pathway, eight differential miRNAs (hsa-miR-23b-3p, -23a-3p, -15b-5p, -24-3p, -4734, -1246, -7641, and -3197) and 39 target genes (e.g., AKT1, AKT2, PIK3CB, PIK3CG, PIK3R1, PIK3CA, and PIK3CD) were involved, as well as some ceRNAs. Besides, for CSC property-related signaling pathways, six miRNAs (hsa-miR-1246, -15b-5p, -30b-5p, -3197, -4734, and -7110-5p) were dramatically enriched in Hedgehog, Notch, and Wnt signaling pathways via regulating 108 target genes (e.g., DVL1, DVL3, WNT3A, and WNT5A). The mTOR and CSC property-associated signaling pathways may be important oncogenic molecular mechanisms in CD133+ A549 cells.




Similar content being viewed by others
References
Mehan MR, Williams SA, Siegfried JM, Bigbee WL, Weissfeld JL, Wilson DO, et al. Validation of a blood protein signature for non-small cell lung cancer. Clinical proteomics. 2014;11:32.
Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90.
Bhattarai B, Schmidt MF, Ghosh M, Sinha Ray A, Manhas S, Oke V, et al. Lung cancer with skin and breast metastasis: a case report and literature review. Case reports in pulmonology. 2015;2015:136970.
Pardal R, Clarke MF, Morrison SJ. Applying the principles of stem-cell biology to cancer. Nat Rev Cancer. 2003;3:895–902.
Clarke MF, Dick JE, Dirks PB, Eaves CJ, Jamieson CH, Jones DL, et al. Cancer stem cells—perspectives on current status and future directions: Aacr workshop on cancer stem cells. Cancer Res. 2006;66:9339–44.
Eramo A, Lotti F, Sette G, Pilozzi E, Biffoni M, Di Virgilio A, et al. Identification and expansion of the tumorigenic lung cancer stem cell population. Cell Death Differ. 2007;15:504–14.
Pine SR, Marshall B, Varticovski L. Lung cancer stem cells. Dis Markers. 2008;24:257–66.
Wu Y, Wu PY. Cd133 as a marker for cancer stem cells: progresses and concerns. Stem Cells Dev. 2009;18:1127–34.
Bertolini G, Roz L, Perego P, Tortoreto M, Fontanella E, Gatti L, et al. Highly tumorigenic lung cancer cd133+ cells display stem-like features and are spared by cisplatin treatment. Proc Natl Acad Sci U S A. 2009;106:16281–6.
Meng X, Li M, Wang X, Wang Y, Ma D. Both cd133+ and cd133- subpopulations of a549 and h446 cells contain cancer-initiating cells. Cancer Sci. 2009;100:1040–6.
Cai Z, Ke J, He X, Yuan R, Chen Y, Wu X, et al. Significance of mtor signaling and its inhibitor against cancer stem-like cells in colorectal cancer. Ann Surg Oncol. 2014;21:179–88.
Papadimitrakopoulou V. Development of pi3k/akt/mtor pathway inhibitors and their application in personalized therapy for non-small-cell lung cancer. J Thorac Oncol. 2012;7:1315–26.
Bao B, Ali S, Ahmad A, Li Y, Banerjee S, Kong D, et al. Differentially expressed mirnas in cancer stem like cells: markers for tumor cell aggressiveness of pancreatic cancer. Stem Cells Dev. 2014;23:1947–58.
Garofalo M, Croce CM. Role of micrornas in maintaining cancer stem cells. Adv Drug Deliv Rev. 2015;81:53–61.
Eades G, Zhang Y-S, Li Q-L, Xia J-X, Yao Y, Zhou Q. Long non-coding rnas in stem cells and cancer. World journal of clinical oncology. 2014;5:134.
Nie F-Q, Zhu Q, Xu T-P, Zou Y-F, Xie M, Sun M, Xia R, Lu K-H. Long non-coding rna mvih indicates a poor prognosis for non-small cell lung cancer and promotes cell proliferation and invasion. Tumor Biol 2014:1–8.
van der Meer AD, Vermeul K, Poot AA, Feijen J, Vermes I. A microfluidic wound-healing assay for quantifying endothelial cell migration. Am J Physiol Heart Circ Physiol. 2010;298:H719–25.
Gao X, Gulari E, Zhou X. In situ synthesis of oligonucleotide microarrays. Biopolymers. 2004;73:579–96.
Bolstad BM, Irizarry RA, Åstrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–93.
Wang X. Mirdb: a microrna target prediction and functional annotation database with a wiki interface. RNA. 2008;14:1012–7.
Garcia DM, Baek D, Shin C, Bell GW, Grimson A, Bartel DP. Weak seed-pairing stability and high target-site abundance decrease the proficiency of lsy-6 and other micrornas. Nat Struct Mol Biol. 2011;18:1139–46.
Von Mering C, Huynen M, Jaeggi D, Schmidt S, Bork P, Snel B. String: a database of predicted functional associations between proteins. Nucleic Acids Res. 2003;31:258–61.
Chang L, Graham P, Hao J, Ni J, Bucci J, Cozzi P, et al. Acquisition of epithelial–mesenchymal transition and cancer stem cell phenotypes is associated with activation of the pi3k/akt/mtor pathway in prostate cancer radioresistance. Cell Death Dis. 2013;4:e875.
Kohl M, Wiese S, Warscheid B. Cytoscape: software for visualization and analysis of biological networks; Data mining in proteomics, Springer. 2011: 291–303.
Li J-H, Liu S, Zhou H, Qu L-H, Yang J-H. Starbase v2. 0: decoding mirna-cerna, mirna-ncrna and protein–rna interaction networks from large-scale clip-seq data. Nucleic Acids Res. 2014;42:D92–7.
Takebe N, Harris PJ, Warren RQ, Ivy SP. Targeting cancer stem cells by inhibiting wnt, notch, and hedgehog pathways. Nat Rev Clin Oncol. 2011;8:97–106.
van Roy F. Beyond e-cadherin: roles of other cadherin superfamily members in cancer. Nat Rev Cancer. 2014;14:121–34.
Zhong K, Chen K, Han L, Li B. Microrna-30b/c inhibits non-small cell lung cancer cell proliferation by targeting rab18. BMC Cancer. 2014;14:703.
Kim J, Lim NJ, Jang SG, Kim HK, Lee GK. Mir-592 and mir-552 can distinguish between primary lung adenocarcinoma and colorectal cancer metastases in the lung. Anticancer Res. 2014;34:2297–302.
Li B, Sun M, Gao F, Liu W, Yang Y, Liu H, et al. Up-regulated expression of mir-23a/b targeted the pro-apoptotic fas in radiation-induced thymic lymphoma. Chem Biol Interact. 2013;32:1729–40.
Kim KM, Heo DR, Lee J, Park JS, Baek MG, Yi JM, et al. 5,3′-Dihydroxy-6,7,4′-trimethoxyflavanone exerts its anticancer and antiangiogenesis effects through regulation of the akt/mtor signaling pathway in human lung cancer cells. Chem Biol Interact. 2015;225:32–9.
Takeshita N, Hoshino I, Mori M, Akutsu Y, Hanari N, Yoneyama Y, et al. Serum microrna expression profile: Mir-1246 as a novel diagnostic and prognostic biomarker for oesophageal squamous cell carcinoma. Br J Cancer. 2013;108:644–52.
Zhu LH, Liu T, Tang H, Tian RQ, Su C, Liu M, et al. Microrna-23a promotes the growth of gastric adenocarcinoma cell line mgc803 and downregulates interleukin-6 receptor. FEBS J. 2010;277:3726–34.
Huang S, He X, Ding J, Liang L, Zhao Y, Zhang Z, et al. Upregulation of mir-23a approximately 27a approximately 24 decreases transforming growth factor-beta-induced tumor-suppressive activities in human hepatocellular carcinoma cells. Int J Cancer. 2008;123:972–8.
Cao M, Seike M, Soeno C, Mizutani H, Kitamura K, Minegishi Y, et al. Mir-23a regulates tgf-β-induced epithelial-mesenchymal transition by targeting e-cadherin in lung cancer cells. Int J Oncol. 2012;41:869–75.
Bjornsti M-A, Houghton PJ. The tor pathway: a target for cancer therapy. Nat Rev Cancer. 2004;4:335–48.
Morgensztern D, McLeod HL. Pi3k/akt/mtor pathway as a target for cancer therapy. Anticancer Drugs. 2005;16:797–803.
Esteller M. Non-coding rnas in human disease. Nat Rev Genet. 2011;12:861–74.
Huang K-C, Rao PH, Lau CC, Heard E, Ng S-K, Brown C, et al. Relationship of xist expression and responses of ovarian cancer to chemotherapy 1 this work was partly supported by nih grants ca70216 and gm 59920 (to sw. N.). 1. Mol Cancer Ther. 2002;1:769–76.
Weakley SM, Wang H, Yao Q, Chen C. Expression and function of a large non-coding rna gene xist in human cancer. World J Surg. 2011;35:1751–6.
Rosso SB, Sussman D, Wynshaw-Boris A, Salinas PC. Wnt signaling through dishevelled, rac and jnk regulates dendritic development. Nat Neurosci. 2004;8:34–42.
Reya T, Clevers H. Wnt signalling in stem cells and cancer. Nature. 2005;434:843–50.
Takahashi-Yanaga F, Kahn M. Targeting wnt signaling: can we safely eradicate cancer stem cells? Clin Cancer Res. 2010;16:3153–62.
Yang Y-P, Chien Y, Chiou G-Y, Cherng J-Y, Wang M-L, Lo W-L, et al. Inhibition of cancer stem cell-like properties and reduced chemoradioresistance of glioblastoma using microrna145 with cationic polyurethane-short branch pei. Biomaterials. 2012;33:1462–76.
Acknowledgments
This study was supported by “334” High-tech Talent Training Program of Nanjing Military Command and Natural Science Foundation of Zhejiang Province of China (LY12H16001).
Conflicts of interest
None
Author information
Authors and Affiliations
Corresponding authors
Additional information
Highlights
1. CD133+ A549 cells showed greater carcinogenic potential than CD133− subpopulation.
2. Totally, 14 differentially expressed miRNAs were identified in CD133+ A549 cells.
3. Totally, 119 signaling pathways were enriched by 14 differentially expressed miRNAs.
4. The mTOR signaling pathway was enriched by eight dysregulated miRNAs via 39 targets.
5. Stem cell property-related pathways were enriched by six dysregulated miRNAs.
Qing-yong Chen, De-min Jiao and Ya Zhu contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Fig. 1
CD133 expression checked by flow cytometry. a, CD133 expression of harvested cells before magnetic activated cell sorting; b, CD133 expression of CD133+ fractions after magnetic activated cell sorting. (GIF 15 kb)
Supplementary Fig. 2
Network of differentially expressed miRNA and CD133 (PROM1). Red circle represents target gene, and green triangle represents differentially expressed miRNA. miRNA, microRNA. (GIF 46 kb)
Rights and permissions
About this article
Cite this article
Chen, Qy., Jiao, Dm., Zhu, Y. et al. Identification of carcinogenic potential-associated molecular mechanisms in CD133+ A549 cells based on microRNA profiles. Tumor Biol. 37, 521–530 (2016). https://doi.org/10.1007/s13277-015-3675-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-015-3675-9