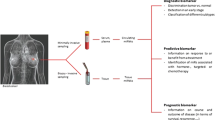
Abstract
Over the past decade, microRNAs (miRNAs) have become a new paradigm of gene regulation. miRNAs are involved in a wide array of carcinogenic processes. Indeed, increasing evidence has shown the importance of miRNAs in cancer, suggesting their possible use as diagnostic, predictive and prognostic biomarkers, leading to miRNA-based anti-cancer therapies, either alone or in combination with current targeted therapies, with the goal of improving cancer treatment responses and increasing cure rates. The advantage of using a miRNA approach is based on the ability to concurrently target multiple effectors of pathways involved in cell proliferation, migration and survival. This review sheds new light on miRNA regulation of genes that play critical roles in the process of malignant transformation and tumour metastasis, the dysregulation of miRNA expression in cancer development and the development of miRNA-based diagnostics and therapeutics.

Similar content being viewed by others
References
Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA. 2013;63:11–30.
Mukherji S, Ebert MS, Zheng GX, Tsang JS, Sharp PA, van Oudenaarden A. MicroRNAs can generate thresholds in target gene expression. Nat Genet. 2011;43:854–9.
Dvinge H, Git A, Gräf S, Salmon-Divon M, Curtis C, Sottoriva A, et al. The shaping and functional consequences of the microRNA landscape in breast cancer. Nature. 2013;497:378–82.
Shen J, Xia W, Khotskaya YB, Huo L, Nakanishi K, Lim S-O, et al. EGFR modulates microRNA maturation in response to hypoxia through phosphorylation of AGO2. Nature. 2013;497:383–7.
Song SJ, Poliseno L, Song MS, Ala U, Webster K, Ng C, et al. MicroRNA-antagonism regulates breast cancer stemness and metastasis via TET-family-dependent chromatin remodeling. Cell. 2013;154:311–24.
Luo Q, Li X, Gao Y, Long Y, Chen L, Huang Y, et al. MiRNA-497 regulates cell growth and invasion by targeting cyclin E1 in breast cancer. Cancer Cell Int. 2013;13:95.
Cui W, Zhang S, Shan C, Zhou L, Zhou Z. MicroRNA-133a regulates the cell cycle and proliferation of breast cancer cells by targeting epidermal growth factor receptor through the EGFR/Akt signaling pathway. FEBS J. 2013;280:3962–74.
Di Leva G, Piovan C, Gasparini P, Ngankeu A, Taccioli C, Briskin D, et al. Estrogen mediated-activation of miR-191/425 cluster modulates tumorigenicity of breast cancer cells depending on estrogen receptor status. PLoS Genet. 2013;9:e1003311.
Leivonen S-K, Sahlberg KK, Mäkelä R, Due EU, Kallioniemi O, Børresen-Dale A-L, et al. High-throughput screens identify microRNAs essential for HER2 positive breast cancer cell growth. Mol Oncol. 2014;8:93–104.
Nassirpour R, Mehta PP, Baxi SM. Yin M-J: miR-221 promotes tumorigenesis in human triple negative breast cancer cells. PLoS One. 2013;8:e62170.
Tanic M, Yanowsky K, Rodriguez-Antona C, Andrés R, Márquez-Rodas I, Osorio A, et al. Deregulated miRNAs in hereditary breast cancer revealed a role for miR-30c in regulating KRAS oncogene. PLoS One. 2012;7:e38847.
Körner C, Keklikoglou I, Bender C, Wörner A, Münstermann E, Wiemann S. MicroRNA-31 sensitizes human breast cells to apoptosis by direct targeting of protein kinase C epsilon (PKCepsilon). J Biol Chem. 2013;288:8750–61.
Anaya-Ruiz M, Cebada J, Delgado-López G, Luisa M. miR-153 silencing induces apoptosis in the MDA-MB-231 breast cancer cell line. Asian Pac J Cancer Prev. 2013;14:2983–6.
Gao J, Li L, Wu M, Liu M, Xie X, Guo J, et al. MiR-26a inhibits proliferation and migration of breast cancer through repression of MCL-1. PLoS One. 2013;8:e65138.
Hwang MS, Yu N, Stinson SY, Yue P, Newman RJ, Allan BB, et al. miR-221/222 targets adiponectin receptor 1 to promote the epithelial-to-mesenchymal transition in breast cancer. PLoS One. 2013;8:e66502.
Arora H, Qureshi R, Park W-Y. miR-506 regulates epithelial mesenchymal transition in breast cancer cell lines. PLoS One. 2013;8:e64273.
Tavazoie SF, Alarcón C, Oskarsson T, Padua D, Wang Q, Bos PD, et al. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147–52.
Zhang N, Wang X, Huo Q, Sun M, Cai C, Liu Z, Hu G, Yang Q: MicroRNA-30a suppresses breast tumor growth and metastasis by targeting metadherin. Oncogene 2013
Li L-Z, Zhang CZ, Liu L-L, Yi C, Lu S-X, Zhou X, et al. Yun J-P: miR-720 inhibits tumor invasion and migration in breast cancer by targeting TWIST1. Carcinogenesis. 2014;35:469–78.
Yang J, Zhang Z, Chen C, Liu Y, Si Q, Chuang T, et al. MicroRNA-19a-3p inhibits breast cancer progression and metastasis by inducing macrophage polarization through downregulated expression of Fra-1 proto-oncogene. Oncogene. 2014;33:3014–23.
Cai J, Guan H, Fang L, Yang Y, Zhu X, Yuan J, et al. MicroRNA-374a activates Wnt/β-catenin signaling to promote breast cancer metastasis. J Clin Invest. 2013;123:566.
Taylor MA, Sossey-Alaoui K, Thompson CL, Danielpour D, Schiemann WP. TGF-β upregulates miR-181a expression to promote breast cancer metastasis. J Clin Invest. 2013;123:150.
Yu S-J, Hu J-Y, Kuang X-Y, Luo J-M, Hou Y-F, Di G-H, et al. MicroRNA-200a promotes anoikis resistance and metastasis by targeting YAP1 in human breast cancer. Clin Cancer Res. 2013;19:1389–99.
Lei R, Tang J, Zhuang X, Deng R, Li G, Yu J, et al. Suppression of MIM by microRNA-182 activates RhoA and promotes breast cancer metastasis. Oncogene. 2014;33:1287–96.
Okuda H, Xing F, Pandey PR, Sharma S, Watabe M, Pai SK, et al. Liu Y: miR-7 suppresses brain metastasis of breast cancer stem-like cells by modulating KLF4. Cancer Res. 2013;73:1434–44.
Hu F, Meng X, Tong Q, Liang L, Xiang R, Zhu T, et al. BMP-6 inhibits cell proliferation by targeting microRNA-192 in breast cancer. Biochim Biophys Acta. 1832;2013:2379–90.
Chou J, Lin JH, Brenot A, Kim J-W, Provot S, Werb Z. GATA3 suppresses metastasis and modulates the tumour microenvironment by regulating microRNA-29b expression. Nat Cell Biol. 2013;15:201–13.
Zhu N, Zhang D, Xie H, Zhou Z, Chen H, Hu T, et al. Endothelial-specific intron-derived miR-126 is down-regulated in human breast cancer and targets both VEGFA and PIK3R2. Mol Cell Biochem. 2011;351:157–64.
Siragam V, Rutnam ZJ, Yang W, Fang L, Luo L, Yang X, et al. MicroRNA miR-98 inhibits tumor angiogenesis and invasion by targeting activin receptor-like kinase-4 and matrix metalloproteinase-11. Oncotarget. 2012;3:1370–85.
Zou C, Xu Q, Mao F, Li D, Bian C, Liu L-Z, et al. MiR-145 inhibits tumor angiogenesis and growth by N-RAS and VEGF. Cell Cycle. 2012;11:2137–45.
Kong W, He L, Richards E, Challa S, Xu C, Permuth-Wey J, et al. Upregulation of miRNA-155 promotes tumour angiogenesis by targeting VHL and is associated with poor prognosis and triple-negative breast cancer. Oncogene. 2014;33:679–89.
Xu Q, Jiang Y, Yin Y, Li Q, He J, Jing Y, et al. A regulatory circuit of miR-148a/152 and DNMT1 in modulating cell transformation and tumor angiogenesis through IGF-IR and IRS1. J Mol Cell Biol. 2013;5:3–13.
He T, Qi F, Jia L, Wang S, Song N, Guo L, et al. MicroRNA-542-3p inhibits tumor angiogenesis by targeting angiopoietin-2. J Pathol. 2014;232:499–508.
Yang R, Schlehe B, Hemminki K, Sutter C, Bugert P, Wappenschmidt B, et al. A genetic variant in the pre-miR-27a oncogene is associated with a reduced familial breast cancer risk. Breast Cancer Res Treat. 2010;121:693–702.
Zhong S, Chen Z, Xu J, Li W, Zhao J. Pre-mir-27a rs895819 polymorphism and cancer risk: a meta-analysis. Mol Biol Rep. 2013;40:3181–6.
Lian H, Wang L, Zhang J. Increased risk of breast cancer associated with CC genotype of Has-miR-146a rs2910164 polymorphism in Europeans. PLoS One. 2012;7:e31615.
Fan C, Chen C, Wu D. The association between common genetic variant of microRNA-499 and cancer susceptibility: a meta-analysis. Mol Biol Rep. 2013;40:3389–94.
Wang J, Bi J, Liu X, Li K, Di J, Wang B. Has-miR-146a polymorphism (rs2910164) and cancer risk: a meta-analysis of 19 case-control studies. Mol Biol Rep. 2012;39:4571–9.
Wang J, Wang Q, Liu H, Shao N, Tan B, Zhang G, et al. The association of miR-146a rs2910164 and miR-196a2 rs11614913 polymorphisms with cancer risk: a meta-analysis of 32 studies. Mutagenesis. 2012;27:779–88.
Wang P-Y, Gao Z-H, Jiang Z-H, Li X-X, Jiang B-F, Xie S-Y. The associations of single nucleotide polymorphisms in miR-146a, miR-196a and miR-499 with breast cancer susceptibility. PLoS One. 2013;8:e70656.
Chen J, Qin Z, Jiang Y, Wang Y, He Y, Dai J, et al. Genetic variations in the flanking regions of miR-101-2 are associated with increased risk of breast cancer. PLoS One. 2014;9:e86319.
Guan X, Liu H, Ju J, Li Y, Li P, Wang LE, Brewster AM, Buchholz TA, Arun BK, Wei Q: Genetic variant rs16430 6bp > 0bp at the microRNA-binding site in TYMS and risk of sporadic breast cancer risk in non-Hispanic white women aged ≤55 years. Molecular Carcinogenesis 2013
Gilam A, Edry L, Mamluk-Morag E, Bar-Ilan D, Avivi C, Golan D, et al. Involvement of IGF-1R regulation by miR-515-5p modifies breast cancer risk among BRCA1 carriers. Breast Cancer Res Treat. 2013;138:753–60.
Jiang Y, Qin Z, Hu Z, Guan X, Wang Y, He Y, et al. Genetic variation in a hsa-let-7 binding site in RAD52 is associated with breast cancer susceptibility. Carcinogenesis. 2013;34:689–93.
Jiang Y, Chen J, Wu J, Hu Z, Qin Z, Liu X, et al. Evaluation of genetic variants in microRNA biosynthesis genes and risk of breast cancer in Chinese women. Int J Cancer. 2013;133:2216–24.
Si H, Sun X, Chen Y, Cao Y, Chen S, Wang H, et al. Circulating microRNA-92a and microRNA-21 as novel minimally invasive biomarkers for primary breast cancer. J Cancer Res Clin Oncol. 2013;139:223–9.
Chan M, Liaw CS, Ji SM, Tan HH, Wong CY, Thike AA, et al. Identification of circulating microRNA signatures for breast cancer detection. Clin Cancer Res. 2013;19:4477–87.
Cuk K, Zucknick M, Madhavan D, Schott S, Golatta M, Heil J, et al. Plasma microRNA panel for minimally invasive detection of breast cancer. PLoS One. 2013;8:e76729.
Chen W, Cai F, Zhang B, Barekati Z, Zhong XY. The level of circulating miRNA-10b and miRNA-373 in detecting lymph node metastasis of breast cancer: potential biomarkers. Tumor Biol. 2013;34:455–62.
Kumar S, Keerthana R, Pazhanimuthu A, Perumal P. Overexpression of circulating miRNA-21 and miRNA-146a in plasma samples of breast cancer patients. Indian J Biochem Biophys. 2013;50:210–4.
Zeng RC, Zhang W, Yan XQ, Ye ZQ, Chen ED, Huang DP, et al. Down-regulation of miRNA-30a in human plasma is a novel marker for breast cancer. Med Oncol. 2013;30:1–8.
Ng EK, Li R, Shin VY, Jin HC, Leung CP, Ma ES, et al. Circulating microRNAs as specific biomarkers for breast cancer detection. PLoS One. 2013;8:e53141.
Markou A, Yousef GM, Stathopoulos E, Georgoulias V, Lianidou E. Prognostic significance of metastasis-related microRNAs in early breast cancer patients with a long follow-up. Clin Chem. 2014;60:197–205.
Wang S, Li H, Wang J, Wang D. Expression of microRNA-497 and its prognostic significance in human breast cancer. Diagn Pathol. 2013;8:172.
Falkenberg N, Anastasov N, Rappl K, Braselmann H, Auer G, Walch A, et al. MiR-221/-222 differentiate prognostic groups in advanced breast cancers and influence cell invasion. Br J Cancer. 2013;109:2714–23.
Tang D, Zhang Q, Zhao S, Wang J, Lu K, Song Y, et al. The expression and clinical significance of microRNA-1258 and heparanase in human breast cancer. Clin Biochem. 2013;46:926–32.
Hoppe R, Achinger-Kawecka J, Winter S, Fritz P, Lo W-Y, Schroth W, et al. Increased expression of miR-126 and miR-10a predict prolonged relapse-free time of primary oestrogen receptor-positive breast cancer following tamoxifen treatment. Eur J Cancer. 2013;49:3598–608.
Ma L, Li GZ, Wu ZS, Meng G. Prognostic significance of let-7b expression in breast cancer and correlation to its target gene of BSG expression. Med Oncol. 2014;31:1–5.
Volinia S, Croce CM. Prognostic microRNA/mRNA signature from the integrated analysis of patients with invasive breast cancer. Proc Natl Acad Sci. 2013;110:7413–7.
Bockhorn J, Dalton R, Nwachukwu C, Huang S, Prat A, Yee K, et al. MicroRNA-30c inhibits human breast tumour chemotherapy resistance by regulating TWF1 and IL-11. Nat Commun. 2013;4:1393.
Fang Y, Shen H, Cao Y, Li H, Qin R, Chen Q, et al. Involvement of miR-30c in resistance to doxorubicin by regulating YWHAZ in breast cancer cells. Braz J Med Biol Res. 2014;47:60–9.
Yin J, Zheng G, Jia X, Zhang Z, Zhang W, Song Y, et al. A Bmi1-miRNAs cross-talk modulates chemotherapy response to 5-fluorouracil in breast cancer cells. PLoS One. 2013;8:e73268.
Chen Y, Sun Y, Chen L, Xu X, Zhang X, Wang B, et al. miRNA-200c increases the sensitivity of breast cancer cells to doxorubicin through the suppression of E-cadherin-mediated PTEN/Akt signaling. Mol Med Rep. 2013;7:1579–84.
Zhong S, Li W, Chen Z, Xu J, Zhao J. MiR-222 and miR-29a contribute to the drug-resistance of breast cancer cells. Gene. 2013;531:8–14.
Jiao X, Zhao L, Ma M, Bai X, He M, Yan Y, et al. MiR-181a enhances drug sensitivity in mitoxantone-resistant breast cancer cells by targeting breast cancer resistance protein (BCRP/ABCG2). Breast Cancer Res Treat. 2013;139:717–30.
Hu H, Li S, Cui X, Lv X, Jiao Y, Yu F, et al. The overexpression of hypomethylated miR-663 induces chemotherapy resistance in human breast cancer cells by targeting heparin sulfate proteoglycan 2 (HSPG2). J Biol Chem. 2013;288:10973–85.
Shibahara Y, Miki Y, Onodera Y, Hata S, Chan MS, Yiu CC, et al. Aromatase inhibitor treatment of breast cancer cells increases the expression of let-7f, a microRNA targeting CYP19A1. J Pathol. 2012;227:357–66.
Ward A, Balwierz A, Zhang J, Küblbeck M, Pawitan Y, Hielscher T, et al. Re-expression of microRNA-375 reverses both tamoxifen resistance and accompanying EMT-like properties in breast cancer. Oncogene. 2013;32:1173–82.
He YJ, Wu JZ, Ji MH, Ma T, Qiao EQ, Ma R, et al. Mir‑342 is associated with estrogen receptor‑α expression and response to tamoxifen in breast cancer. Exp Ther Med. 2013;5:813–8.
Lin J, Liu C, Gao F, Mitchel R, Zhao L, Yang Y, et al. miR-200c enhances radiosensitivity of human breast cancer cells. J Cell Biochem. 2013;114:606–15.
Stankevicins L, da Silva APA, dos Passos FV, dos Santos FE, Ribeiro MCM, David MG, et al. MiR-34a is up-regulated in response to low dose, low energy x-ray induced DNA damage in breast cells. Radiat Oncol. 2013;8:231.
Gong C, Yao Y, Wang Y, Liu B, Wu W, Chen J, et al. Up-regulation of miR-21 mediates resistance to trastuzumab therapy for breast cancer. J Biol Chem. 2011;286:19127–37.
Bai WD, Ye XM, Zhang MY, Zhu HY, Xi WJ, Huang X, Zhao J, Gu B, Zheng GX, Yang AG: MiR-200c suppresses TGF-β signaling and counteracts trastuzumab resistance and metastasis by targeting ZNF217 and ZEB1 in breast cancer. International Journal of Cancer 2014
Ye X, Bai W, Zhu H, Zhang X, Chen Y, Wang L, et al. MiR-221 promotes trastuzumab-resistance and metastasis in HER2-positive breast cancers by targeting PTEN. BMB Rep. 2014;47:268–73.
Jung EJ, Santarpia L, Kim J, Esteva FJ, Moretti E, Buzdar AU, et al. Plasma microRNA 210 levels correlate with sensitivity to trastuzumab and tumor presence in breast cancer patients. Cancer. 2012;118:2603–14.
Ye X-M, Zhu H-Y, Bai W-D, Wang T, Wang L, Chen Y, et al. Epigenetic silencing of miR-375 induces trastuzumab resistance in HER2-positive breast cancer by targeting IGF1R. BMC Cancer. 2014;14:134.
Conflicts of interest
None
Author information
Authors and Affiliations
Corresponding author
Additional information
WenCheng Zhang is the first author.
Rights and permissions
About this article
Cite this article
Zhang, W., Liu, J. & Wang, G. The role of microRNAs in human breast cancer progression. Tumor Biol. 35, 6235–6244 (2014). https://doi.org/10.1007/s13277-014-2202-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-014-2202-8