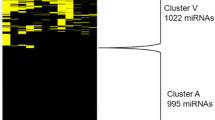
Abstract
The goal of this study was to compare and analyze differentially expressed miRNA and their targets in human chondrosarcoma JJ012 and chondrocytes C 28 cell lines (control) to elucidate deregulation of major signal transduction pathways involved in sarcomagenesis. Total RNA extraction was followed by analyzing RNA quality and integrity. Exiqon human miRNA panel of 743 unique miRNA assays and Illumina microarray HT-12 platform and quantitative reverse transcriptase–PCR verification of targets were performed. The results from human miRNA Exiqon arrays with biological triplicates indicated 28 significant miRNAs (P value ≤0.01). A total 3,045 target genes were derived from the miRWalk database for these 28 miRNAs with 587 common and 2,458 unique target genes. The results of our analyses of the significantly downregulated and upregulated miRNAs in chondrosarcoma cell line indicated the predominant dysregulation of NOTCH, insulin-like growth factor receptor (IGFR), and downstream rat sarcoma, and Src pathways, compared to control. Among the upregulated targets for upregulated miRNAs were the cluster of cancer testis antigen (CTA) genes, located on X chromosome, and their expression was correlated to IGFR pathway activity. Based on our observations, lost miRNA surveillance of NOTCH and IGFR pathways is involved in and leads to sarcomagenesis. We conclude that upregulation of CTA genes is due to hypomethylation that are controlled by epi-miRNAs. We do not preclude the possibility that the upregulated miRNAs, which target CTA genes located in adjacent regions in chromosome X, are epi-miRNAs that influence target gene expression by directly regulating epigenetic processes. Future endeavors will be directed towards understanding the posttranscriptional modifications that affect miRNA expression in sarcomas.


Similar content being viewed by others
References
Mirra J. Bone tumors. Philadelphia: Lea and Febiger; 1989.
Frassica FJ, Unni KK, Beabout JW, Sim FH. Dedifferentiated chondrosarcoma. A report of the clinicopathological features and treatment of seventy-eight cases. J Bone Jt Surg Am. 1986;68:1197–205.
Galoian KA, Temple HT, Galoyan AA. mTORC1 inhibition and ECM–cell adhesion-independent drug resistance via PI3K–AKT and PI3K–RAS–MAPK feedback loops. Tumour Biol. 2012;33(3):885–90.
Galoian KA, Garamszegi N, Garamszegi S, Scully S. Molecular mechanism of tenascin-C action on matrix metalloproteinase-1 invasive potential. Exp Biol Med. 2007;232(4):515–22.
Bustinza-Linares E, Socola F, Ernani V, Miller SA, Trent JC. Extraskeletal myxoid chondrosarcoma with small bowel metastasis causing bowel obstruction. Case Rep Oncol Med. 2012;2012:621025.
Ayalon D, Glaser T, Werner H. Transcriptional regulation of IGF-I receptor gene expression by the PAX3-FKHR oncoprotein. Growth Horm IGF Res. 2001;11(5):289–97. 2001.
Karnieli E, Werner H, Rauscher FJ, Benjamin LE, Leroith D. The IGF-I receptor gene promoter is a molecular target for the Ewing's sarcoma-Wilms' tumor 1 fusion protein. J Biol Chem. 1996;271(32):19304–9.
Finkeltov I, Kuhn S, Glaser T, Idelman G, Wright JJ, Roberts Jr CT, et al. Transcriptional regulation of IGF-I receptor gene expression by novel isoforms of the EWS-WT1 fusion protein. Oncogene. 2002;21(12):1890–8.
Werner H, Idelman G, Rubinstein M, Pattee P, Nagalla SR, Roberts CT. A novel EWS-WT1 gene fusion product in desmoplastic small round cell tumor is a potent transactivator of the insulin-like growth factor-I receptor (IGF-IR) gene. Cancer Lett. 2007;24(1–2):84–90.
Pedone PV, Tirabosco R, Cavazzana AO, Ungaro P, Basso G, Luksch R, et al. Mono- and bi-allelic expression of insulin-like growth factor II gene in human muscle tumors. Hum Mol Genet. 1994;3(7):1117–21.
Scrable H, Cavenee W, Ghavimi F, Lovell M, Morgan K, Sapienza C. A model for embryonal rhabdomyosarcoma tumorigenesis that involves genome imprinting. Proc Natl Acad Sci U S A. 1989;86(19):7480–4.
Visser M, Sijmons C, Bras J, Arceci RJ, Godfried M, Valentijn LJ, et al. Allelotype of pediatric rhabdomyosarcoma. Oncogene. 1997;15(11):1309–14.
Zhan S, Shapiro DN, Helman LJ. Activation of an imprinted allele of the insulin-like growth factor II gene implicated in rhabdomyosarcoma. JClin Inv. 1994;94(1):445–8.
Anderson J, Gordon A, Pritchard-Jones K, Shipley J. Genes, chromosomes, and rhabdomyosarcoma. Gene Chromosome Cancer. 1999;26:275–85.
Sun Y, Gao D, Liu Y, Huang J, Lessnick S, Tanaka S. IGF2 is critical for tumorigenesis by synovial sarcoma oncoprotein SYT-SSX1. Oncogene. 2006;25(7):1042–52.
de Bruijn DR, Allander SV, van Dijk AH, Willemse MP, Thijssen J, van Groningen JJ, et al. The synovial sarcoma-associated SS18-SSX2 fusion protein induces epigenetic gene deregulation. Cancer Res. 2006;66(19):9474–82.
Ho L, Stojanovski A, Whetstone H, Wei QX, Mau E, Wunder JS, et al. Gli2 and p53 cooperate to regulate IGFBP-3- mediated chondrocyte apoptosis in the progression from benign to malignant cartilage tumors. Cancer Cell. 2009;16(2):126–3.
Demicco EG, Lazar AJ. Clinicopathologic considerations: how can we fine tune our approach to sarcoma? Semin Oncol. 2011;38:S3–18.
Eliasz S, Liang S, Chen Y, de Marco MA, Machek O, Skucha S, et al. Notch-1 stimulates survival of lung adenocarcinoma cells during hypoxia by activating the IGF-1R pathway. Oncogene. 2010;29(17):2488–98.
Tanaka M, Setoguchi T, Hirotsu M, Gao H, Sasaki H, Matsunoshita Y, et al. Inhibition of Notch pathway prevents osteosarcoma growth by cell cycle regulation. Br J Cancer. 2009;100(12):1957–65.
Garcia A, Kandel JJ. Notch: a key regulator of tumor angiogenesis and metastasis. Histol Histopathol. 2012;27(2):151–6.
Sjolund J, Johansson M, Manna S, Norin C, Pietras A, Beckman S, et al. Suppression of renal cell carcinoma growth by inhibition of Notch signaling in vitro and in vivo. J Clin Invest. 2008;118:217–28.
Honoki K, Fujii H, Tohma Y, Tsujiuchi T, Kido A, Tsukamoto S, et al. Comparison of gene expression profiling in sarcomas and mesenchymal stem cells identifies tumorigenic pathways in chemically induced rat sarcoma model. Oncol. 2012;2012:909453.
Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–54.
Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6(11):857–6.
Drakaki A, Iliopoulos D. MicroRNA gene networks in oncogenesis. Curr Genomics. 2009 Mar;10(1):35–41.
Place RF, Li LC, Pookot D, Noonan EJ, Dahiya R. MicroRNA-373 induces expression of genes with complementary promoter sequences. Proc Natl Acad Sci U S A. 2008;105(5):1608–13.
Portnoy V, Huang V, Place RF, Li LC. Small RNA and transcriptional upregulation. Wiley Interdiscip Rev RNA. 2011;2(5):748–60.
Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA. 2004;101:2999–3004.
Hatziapostolou M, Polytarchou C, Iliopoulos D. miRNAs link metabolic reprogramming to oncogenesis. Trends Endocrinol Metab. 2013;24(7):361–73.
Dela Cruz F, Matushansky I. MicroRNAs in chromosomal translocation-associated solid tumors: learning from sarcomas. Discov Med. 2011;12(65):307–17.
Dweep H, Sticht C, Pandey P, Gretz N. miRWalk - database: prediction of possible miRNA binding sites by “walking” the genes of 3 genomes. J Biomed Inform. 2011;44:839–7.
Wang D, Qiu C, Zhang H, Wang J, Cui Q, Yin Y. Human MicroRNA oncogenes and tumor suppressors show significantly different biological patterns: from functions to targets. PLoS One. 2010;5(9):e1306.
Bandyopadhyay S, Mitra R, Maulik U, Zhang M. Development of the human cancer microRNA network. Silence. 2010;1:6.
Li Y, Yu X, Lin S, Li X, Zhang S, Song YH. Insulin-like growth factor 1 enhances the migratory capacity of mesenchymal stem cells. Biochem Biophys Res Commun. 2007;356(3):780–4.
Salani B, Passalacqua M, Maffioli S, Briatore L, Hamoudane M, Contini P, et al. IGF-IR internalizes with caveolin-1 and PTRF/Cavin in Hacat Cells. PLoS One. 2010;5(11):e14157.
Weeraratne SD, Amani V, Neiss A, Teider N, Scott DK, Pomeroy ST, et al. miR-34a confers chemosensitivity through modulation of MAGE-A and p53 in medulloblastoma. Neuro Oncol. 2011;13(2):165–75.
Shah MY, Calin GA. MicroRNAs miR-221 and miR-222: a new level of regulation in aggressive breast cancer. Genome Med. 2011;3(8):56.
Solomon A, Mian Y, Ortega-Cava C, Liu V, Gurumurthy CB, Naramura M, et al. Upregulation of the let-7 microRNA with precocious development in lin-12/Notch hypermorphic C. elegans mutants. Dev Biol. 2008;316(2):191–9.
Brueckner B, Stresemann C, Kuner R, Mund C, Musch T, Meister M, et al. The human let-7a-3 locus contains an epigenetically regulated microRNA gene with oncogenic function. Cancer Res. 2007;67(4):1419–23.
Himpe E, Kooijman R. Insulin-like growth factor-I receptor signal transduction and the Janus Kinase/Signal Transducer and Activator of Transcription (JAK-STAT) pathway. Biofactors. 2009;35(1):76–81.
Liu Y, Miao Y, Wang J, Lin X, Wang L, Xu HT, et al. DEC1 is positively associated with the malignant phenotype of invasive breast cancers and negatively correlated with the expressor of claudin 1. Int J Mol med. 2013;31(4):855–60.
Hatekayama N, Kojima T, Iba K, Murata M, Thi MM, Spray DC, et al. IGF regulates tight junction protein claudin 1 during differentiation of osteoblast like MC3T3 E1 cell via MAP kinase pathway. Cell Tissue Res. 2008;334(2):243–54.
Grey A, Chen Q, Callon K, Xu X, Cornish J. Parallel phosphatidylinositol-3 kinase and p42/44 mitogen-activated protein kinase signaling pathways subserve the mitogenic and antiapoptotic actions of insulin-like growth factor I in osteoblastic cells. Endocrinology. 2003;144:4886–93.
Namkung J, Kim M.Y, Woo M.K, Sohn J.H, An S.W and Yeh B, I. Molecular mechanism of RBP4 on the basis of insulin signaling pathway in primary hepatocytes. FASEB J. 2010; 24 (Meeting Abstract Supplement): 88
Harvey AJ, Mao S, Lalancette C, Krawetz SA, Brenner CA. Transcriptional differences between rhesus embryonic stem cells generated from in vitro and in vivo derived embryos. PLoS One. 2012;7(9):e43239.
de Backer O, Arden KC, Boretti M, Vantomme V, de Smet C, Czekay S, et al. Characterization of the GAGE genes that are expressed in various human cancers and in normal testis. Cancer Res. 1999;59(13):3157–65.
Stefan M, Simmons RA, Bertera S, Trucco M, Esni F, Drain P, et al. Global deficits in development, function, and gene expression in the endocrine pancreas in a deletion mouse model of Prader–Willi syndrome. Am J Physiol Endocrinol Metab. 2011;300:E909–22.
Suzuki H, Maruyama R, Yamamoto E, Kai M. DNA methylation and microRNA dysregulation in cancer. Mol Oncol. 2012;6(6):567–78.
Karpf AR, Bai S, James SR, Mohler JL, Wilson EM. Increased expression of androgen receptor coregulator MAGE-11 in prostate cancer by DNA hypomethylation and cyclic AMP. Mol Cancer Res. 2009;7:523–35.
Lim JH, Kim SP, Gabrielson E, Park YB, Park JW, Kwon TK. Activation of human cancer/testis antigen gene, XAGE-1, in tumor cells is correlated with CpG island hypomethylation. Int J Cancer. 2005;116:200–6.
Xiao J, Chen HS, Fei R, Cong X, Wang LP, Wang Y, et al. Expression of MAGE-A1 mRNA is associated with gene hypomethylation in hepatocarcinoma cell lines. J Gastroenterol. 2005;40:716–21.
Fabbri M, Calore F, Paone A, Galli R, Calin GA. Epigenetic regulation of miRNAs in cancer. Adv Exp Med Biol. 2013;754:137–48.
Drakaki A, Iliopoulos D. MicroRNA gene networks in oncogenesis. Curr Genomics. 2009;10(1):35.
Rosenfeld N, Aharonov R, Meiri E, Rosenwald S, Spector Y, Zepeniuk M, et al. Barshack I microRNAs accurately identify cancer tissue origin. Nat Biotechnol. 2008;26(4):462–9.
Elmén J, Lindow M, Schütz S, Lawrence M, Petri A, Obad S, et al. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452(7189):896–9.
Monroig PD, Calin GA. MicroRNA and epigenetics: diagnostic and therapeutic opportunities. Curr Pathobiol Rep. 2013;1(1):43–52.
Martín Liberal J, Lagares-Tena L, Sáinz-Jaspeado M, Mateo-Lozano S, García Del Muro X, Tirado OM. Targeted therapies in sarcomas: challenging the challenge. Sarcoma. 2012;2012:626094.
Acknowledgments
The study was financially supported by the University of Miami Tissue bank research account. We would like to thank Yoslalyma Cardentey and Kathy Slosek from the University of Miami Oncogenomics Core Facility for their help and expertise.
Conflicts of interest
None
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Galoian, K., Guettouche, T., Issac, B. et al. Lost miRNA surveillance of Notch, IGFR pathway—road to sarcomagenesis. Tumor Biol. 35, 483–492 (2014). https://doi.org/10.1007/s13277-013-1068-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-013-1068-5