Abstract
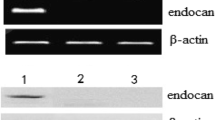
Peptide-based immunotherapy strategies appear promising as an approach to successfully induce an antitumor immune response and prolong survival in patients with various cancers. Protein antigens and their specific epitopes are formulation targets for anti-tumor vaccines. Bioinformatical approaches to predict major histocompatibility complex binding peptides can facilitate the resource-consuming effort of T cell epitope identification. Proliferating cell nuclear antigen including Ki-67 and PCNA, associated with the proliferation process of the cell, seems to be an attractive new target for tumor-specific immunotherapy. In this study, we predicted seven HLA-A*0201-restricted CTL candidate epitope of Ki-67 and eight epitope of PCNA by computer algorithm SYFPEITHI, BIMAS, and IEDB_ANN. Subsequently, biological functions of these peptides were tested by experiments in vitro. We found Ki-67(280–288) (LQGETQLLV) had the strongest binding-affinity with HLA-A*0201. Further study revealed that Ki-67(280–288) increased the frequency of IFN-γ-producing T cells compared to a negative peptide. Because Ki-67 was broadly expressed in most advanced malignant tumors, indicating a potential anti-tumor application in the future.



Similar content being viewed by others
References
Van der Bruggen P, Chomez P, et al. A gene encoding an antigen recognized by cytolytic T lymphocytes on a human melanoma. Science. 1991;254:1643–7.
Purcell AW, McCluskey J, Rossjohn J. More than one reason to rethink the use of peptides in vaccine design. Nat Rev Drug Discov. 2007;6:404–14.
Van der Burg SH, Bijker MS, Welters MJ, Offringa R, Melief CJ. Improved peptide vaccine strategies,creating synthetic artifical infections to maximize immune efficacy. Adv Drug Deliv Rev. 2006;127:159–69.
Noguchi M, Itoh K, Suekane S, Yao A, Suetsugu N, Katagiri K, et al. Phase I trial of patient-oriented vaccination in HLA-A*0201-positive patients with metastatic hormonerefractory prostate cancer. Cancer Sci. 2004;95:77–84.
Wobser M, Keikavoussi P, Kunzmann V, Weininger M, Andersen MH, Becker JC. Complete remission of liver metastasis of pancreyatic cancer under vaccination with a HLA-A*0201 restricted peptide derived from the universal tumor antigen survivin. Cancer Immunol Immunother. 2006;55:1294–8.
Firbas C, Jilma B, Tauber E. Immunogenicity and safety of a novel therapeutic hepatitis C virus (HCV) peptide vaccine: a randomized, placebo controlled trial for dose optimization in 128 healthy subjects. Vaccine. 2006;24:4343–53.
Pietersz GA, Pouniotis DS, Apostolopoalos V. Design of peptide-based vaccines for cancer. Curr Med Chem. 2006;13:1591–607.
Yang HB, Hsu PI, Chan SH, Lee IC, Shin IS, Chow NH. Growth kinetics of colorectal adenoma-carcinoma sequence: an immunohistochemical study of proliferation cell nuclear antigen expression. Hum Pathol. 1996;10:1071–6.
Bai XZ, Masters JR, O’Donoghue N, Kirby R, Pan LX, Young M, et al. Prognostic markers in clinically localised prostate cancer. Int J Oncol. 1999;14:785–91.
SYFPEITHI [http://www.syfpeithi.de/Scripts/MHCServer.dll/EpitopePrediction.htm], Accessed July 28, 2010.
IEDB (ANN)[http://tools.immuneepitope.org/analyze/html/mhc_binding.html], Accessed July 28, 2010.
Nijman HW, Houbiers JG, Vierboom MP, van der Burg SH, Drijfhout JW, D’Amaro J, et al. IdentiWcation of peptide sequences that potentially trigger HLA-A*0201.1-restricted cytotoxic T lymphocytes. Eur J Immunol. 1993;23:1215–9.
Andersen MH, Pedersen LO, Becker JC, Straten P. Identification of a cytotoxic T lymphocyte response to the apoptosis inhibitor protein survivin in cancer patients. Cancer Res. 2000;61:869–72.
van der Bruggen P, Traversari C, Chomez P, Lurquin C, De Plaen E, Van den Eynde B, et al. A gene encoding an antigen recognized by cytolytic T lymphocytes on a human melanoma. Science. 1991;254:1643–7.
Brusic V, Bajic VB, Petrovsky N. Computational methods for prediction of T-cell epitopes—a framework for modelling, testing, and applications. Methods. 2004;34:436–43.
De Groot AS, Moise L. Prediction of immunogenicity for therapeutic proteins: state of the art. Curr Opin Drug Discov Dev. 2007;10:332–40.
Claus Lundegaard, Kasper Lamberth, Mikkel Harndahl, Søren Buus, Ole Lund1and Morten Nielsen. NetMHC-3.0: accurate web accessible predictions of human, mouse and monkey MHC class I affinities for peptides of length 8–11. Nucleic Acids Res. 2008;36:509–12.
Lim JS, Kim S, Lee HG, Lee KY, Kown TJ, Kim K. Selection of peptides that bind to the HLA-A*0201.1 molecule by molecular modelling. Mol Immunol. 1996;33:221–30.
Lin HH, Ray S, Tongchusak S, Reinherz EL, Brusic V. Evaluation of MHC class I peptide binding prediction servers: applications for vaccine research. BMC Immunol. 2008;9:8.
Yu K, Petrovsky N, Schonbach C, Koh JY, Brusic V. Methods for prediction of peptide binding to MHC molecules: a comparative study. Mol Med (Camb, Mass). 2002;8:137–48.
Trost B, Bickis M, Kusalik A. Strength in numbers: achieving greater accuracy in MHC-I binding prediction by combining the results from multiple prediction tools. Immunome Res. 2007;3:5.
Apostolopoulos V. Peptide-based vaccines for cancer: are we choosing the right peptides? Expert Rev Vaccines. 2009;8:259–60.
Buteau C, Markovic SN, Celis E. Challenges in the development of effective peptide vaccines for cancer. Mayo Clin Proc. 2002;77:339–49.
Shreya Kanodia W, Kast M. peptide-based vaccines for cancer: realizing their potential. Expert Rev Vaccines. 2008;7:1533–45.
Acknowledgements
This project is supported by grants from Health Departmental of Jiangsu province (No. Z200903), National Science Foundation of China (No.30972976) and the Program for New Century Excellent Talents in University (NCET-08-0700).
Author information
Authors and Affiliations
Corresponding author
Additional information
Wei Xu and Hui-Zhong Li contributed equally to this work.
Rights and permissions
About this article
Cite this article
Xu, W., Li, HZ., Liu, JJ. et al. Identification of HLA-A*0201-restricted cytotoxic T lymphocyte epitope from proliferating cell nuclear antigen. Tumor Biol. 32, 63–69 (2011). https://doi.org/10.1007/s13277-010-0098-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-010-0098-5