Abstract
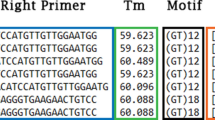
High-resolution melting (HRM) analysis is an emerging technology to screen microsatellites for polymorphism. A potential issue surrounding this method is that amplicon sizes for HRM should typically be short (80–100 bp) for highest sensitivity to reveal polymorphism via the presence of two peaks in the curve of the derivative of fluorescence over temperature (dF/dT). In contrast, microsatellite amplicons are typically 100–400 bp. Therefore, we compared HRM analysis melting temperature range (ΔTm) and multiple dF/dT peaks for predicting microsatellite polymorphism. We assessed polymorphism at 27 microsatellite loci, with estimated lengths of 122–321 bp, in Providence Petrel (Pterodroma solandri). We validated HRM assessment using traditional capillary electrophoresis (CE). While 100 % of loci exhibiting multiple peaks in the dF/dT curve were confirmed as polymorphic by CE, 16 % improvement in sensitivity (83 vs. 67 %) was achieved by using ΔTm, and 25 % (92 vs. 67 %) by using ΔTm in addition to multiple dF/dT peaks. We suggest HRM melting temperature range as new predictor of polymorphism that can be used to rapidly assess microsatellites polymorphism.




Similar content being viewed by others
References
Andersen P, Jespersgaard C, Vuust J, Christiansen M, Larsen L (2003) Capillary electrophoresis-based single strand DNA conformation analysis in high-throughput mutation screening. Hum Mutat 21:455–465
Arthofer W, Steiner F, Schlick-Steiner B (2011) Rapid and cost effective screening of newly identified microsatellite loci by high-resolution melting analysis. Mol Genet Genomics 286:225–235
Bachtrog D, Agis G, Imhof M, Schlotterer C (2000) Microsatellite variability differs between dinucleotide repeat motifs-evidence from Drosophila melanogaster. Mol Biol Evol 17:1277–1285
Brinkmann B, Klintschar M, Neuhuber F, Huhne J, Rolf B (1998) Mutation rate in human microsatellites: influence of the structure and length of the tandem repeat. Am J Hum Genet 62:1408–1415
Brown RM, Jordan WC (2009) Characterization of polymorphic microsatellite loci from Round Island petrels (Pterodroma arminjoniana) and their utility in other seabird species. J Ornithol 150:925–929
Burg TM (1999) Isolation and characterization of microsatellites in albatrosses. Mol Ecol 8:338–341
Crawford A, Kappes S, Paterson K, deGotari M, Dodds K, Freking B, Stone R, Beattie C (1998) Microsatellite evolution: testing the ascertainment biais hypothesis. J Mol Evol 46:256–260
Distefano G, Caruso M, La Malfa S, Gentile A, Wu SB (2012) High resolution melting analysis is a more sensitive and effective alternative to gel-based platforms in analysis of SSR—an example in Citrus. PLoS ONE 7:e4402
Dubois MP, Jarne P, Jouventin P (2005) Ten polymorphic microsatellite markers in the wandering albatross Diomedea exulans. Mol Ecol Notes 5:905–907
Gardner M, Fitch A, Bertozzi T, Lowe A (2011) Rise of the machines—recommendations for ecologists when using next generation sequencing for microsatellite development. Mol Ecol Resour 11:1093–1101
Given AD, Mills JA, Baker AJ (2002) Isolation of polymorphic microsatellite loci from the red-billed gull (Larus novaehollandiae scopulinus) and amplification in related species. Mol Ecol Notes 4:416–418
Guichoux E, Lagache L, Wagner S, Chaumeil P, Leger P, Lepais O, Lepoittevin C, Malausa T, Revardel E, Salin F et al (2011) Current trends in microsatellite genotyping. Mol Ecol Resour 11:591–611
Gundry C, Vandersteen J, Reed G, Pryor R, Chen J, Wittwer C (2003) Amplicon melting analysis with labeled primers: a closed-tube method for differentiating homozygotes and heterozygotes. Clin Chem 49:396–406
Gundry CN, Dobrowolski SF, Martin YR, Robbins TC, Nay LM, Boyd N, Coyne T, Wall MD, Wittwer CT, Teng DHF (2008) Base-pair neutral homozygotes can be discriminated by calibrated high-resolution melting of small amplicons. Nucleic Acids Resour 36:3401–3408
Herrmann M, Durtschi J, Bromley L, Wittwer C, Voelkerding K (2006) Amplicon DNA melting analysis for mutation scanning and genotyping: cross-platform comparison of instruments and dyes. Clin Chem 53:494–503
Hughes C, Queller D (1993) Detection of highly polymorphic microsatellite loci in a species with little allozyme polymorphism. Mol Ecol 2:131–137
IUCN (2011) IUCN Red list of threatened species. IUCN
Kupper C, Horsburgh GJ, Dawson DA, Ffrench-Constant R, Szekely T, Burke T (2007) Characterization of 36 polymorphic microsatellite loci in the Kentish plover (Charadrius alexandrinus) including two sex-linked loci and their amplification in four other Charadrius species. Mol Ecol Notes 7:35–39
Lessa E, Applebaum G (1993) Screening techniques for detecting allelic variation in DNA sequences. Mol Ecol 2:119–129
Liew M, Pryor R, Palais R, Meadows C, Erali M, Lyon E, Wittwer C (2004) Genotyping of single-nucleotide polymorphisms by high-resolution melting of small amplicons. Clin Chem 50:1156–1164
Mackay JF, Wright CD, Bonfiglioli RG (2008) A new approach to varietal identification in plants by microsatellite high resolution melting analysis: application to the verification of grapevine and olive cultivars. Plant Methods 4:8
Muleo R, Colao MC, Miano D, Cirilli M, Intrieri M, Baldoni L, Rugini E (2009) Mutation scanning and genotyping by high-resolution DNA melting analysis in olive germplasm. Genome 52:252–260
Reed G, Wittwer C (2004) Sensitivity and specificity of single-nucleotide polymorphism scanning by high-resolution melting analysis. Clin Chem 50:1748–1754
Rozen S, Skaletsky H (2000) Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132:365–386
Schuelke M (2000) An economic method for the fluorescent labeling of PCR fragments. Nat Biotechnol 18:233–236
Seipp M, Durtschi J, Liew M, Williams J, Damjanovich K, Pont-Kingdon G, Lyon E, Voelkerding KV, Wittwer CT (2007) Unlabeled oligonucleotides as internal temperature controls for genotyping by amplicon melting. J Mol Diagn 9:284–289
Smith BL, Lu CP, Bremer JRA (2010) High-resolution melting analysis (HRMA): a highly sensitive inexpensive genotyping alternative for population studies. Mol Ecol Resour 10:193–196
Sun Z, Gomez-Diaz E, Bailie A, Friesen V (2009) Isolation and characterization of microsatellite loci for storm-petrels. Mol Ecol Resour 9:913–915
Techow NMSM, O’ryan C (2004) Characterization of microsatellite loci in White-chinned petrel (Procellaria aequinoctialis) and cross-amplification in six other procellariiform species. Mol Ecol Notes 4:33–35
Tindall E, Peterson D, Woodbridge P, Schinapy K, Hayes V (2009) Assessing high-resolution melt curve analysis for accurate detection of gene variants in complex DNA fragments. Hum Mutat 30:876–883
Welch AJ, Fleischer RC (2011) Polymorphic microsatellite markers for the endangered Hawaiian petrel (Pterodroma sandwichensis). Conserv Genet Resour 3:581–584
Wittwer CT, Reed GH, Gundry CN, Vandersteen JG, Pryor RJ (2003) High-resolution genotyping by amplicon melting analysis using LCGreen. Clin Chem 49:853–860
Xiao D, Kelleher C, Howard-William E, Meade C (2012) Rapid identification of chloroplast haplotypes using high resolution melting analysis. Mol Ecol Resour 12:894–908
Acknowledgements
The Seaworld Research and Rescue Foundation Inc (Grant SWR/4/2011) supported this work.
Author contributions
Anicee Lombal and Theodore Wenner constructed the manuscript and collected the molecular data. Anicee Lombal performed statistical analyses. Christopher Burridge designed the study and contributed to the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there are no conflicts of interest regarding this study.
Rights and permissions
About this article
Cite this article
Lombal, A.J., Wenner, T.J. & Burridge, C.P. Assessment of high-resolution melting (HRM) profiles as predictors of microsatellite variation: an example in Providence Petrel (Pterodroma solandri). Genes Genom 37, 977–983 (2015). https://doi.org/10.1007/s13258-015-0327-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13258-015-0327-9