Abstract
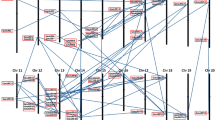
Mungbean is one of the major crops grown in South, East and Southeast Asia because of a high quality of amino acid profile; however, its asynchronous flowering time makes difficult to harvest at a time. Synchronization of flowering time is important to reduce labor costs for harvesting. With the availability of next generation sequencing data of mungbean, we approached a strategy of comparative genomics to identify mungbean homologous counterpart of A. thaliana genes that are known to be involved in flowering pathways, followed by a comparative soybean quantitative trait loci (QTL) analysis of the putative mungbean flowering-related genes. Co-localization of mungbean QTL associated with days to first flower day (FLD) was also identified using the EST-SSR markers from a previous study. Additionally, based on the mungbean transcriptome data with a distinct flowering stage of R2, FPKM (Fragments Per Kilobase per Million mapped reads) expression analysis of all the genes found in paralogous synteny blocks was conducted to examine expression patterns of the genes that have undergone a whole genome duplication event. Our results indicate that the paralogous flowering genes along with other genes within a same synteny block have evolved together at the macro-synteny scale. This study provides insights into mungbean flowering genes, in which they can be used as tools in order to improve flowering synchronization and to increase yield.



Similar content being viewed by others
References
Ali O (2011) Varieties and production technologies of mungbean for R-W-mungbean pattern. IMIS CIMMYT Web. http://imis.cimmyt.org/confluence/download/attachments/26247240/Mung+Production.ppt?version=1. Accessed 7 May 2014
Blanc G, Wolfe KH (2004) Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell 16:1679–1691
Chen H-M et al (2013) The major quantitative trait locus for mungbean yellow mosaic Indian virus resistance is tightly linked in repulsion phase to the major bruchid resistance locus in a cross between mungbean [Vigna radiata (L.) Wilczek] and its wild relative Vigna radiata ssp. sublobata. Euphytica 192:205–216
Cober ER, Tanner JW, Voldeng HD (1999) Soybean photoperiod-sensitivity loci respond differentially to light quality. Crop Sci 36:606–610
Cusack BP, Wolfe KH (2007) When gene marriages don’t work out: divorce by subfunctionalization. Trends Genet 23:270–272
Doebley J, Gaut B, Smith B (2006) The molecular genetics of crop domestication. Cell 127:1309–1321
Edger PP, Pires JC (2009) Gene and genome duplications: the impact of dosage sensitivity on the fate of nuclear genes. Chromosome Res 17:699–717
Fornara F, Montaigu AD, Coupland G (2010) Snapshot: control of flowering in Arabidopsis. Cell 141:550
Henrichsen CN, Chaignat E, Reymond A (2009) Copy number variants, diseases and gene expression. Hum Mol Genet 18:R1–R8
Huang X, Feng Q, Qian Q, Zaho Q, Wang L, Wang A, Guan J, Fan D et al (2009) High-throughput genotyping by whole-genome resequencing. Genome Res 19:1068–1076
Isemura T, Kaga A, Tabata S, Somta P, Srinives P et al (2012) Construction of a Genetic Linkage Map and Genetic Analysis of Domestication Related Traits in Mungbean (Vigna radiata). PLoS One 7:e41304
Jain HK, Mehra KL (1980) Evaluation, adaptation, relationship and cases of the species of Vigna cultivation in Asia. In: Summerfield RJ, Butnting AH (eds) Advances in legume sciences. Royal Botanical Gardens, UK, pp 459–468
Jung C-H, Seo Y-H, Seo PJ, Reyes JL, Bhalla PL (2012) Comparative genomic analysis of soybean flowering genes. PLoS One 7:e38250
Kajonphol T, Sangsiri C, Somta P, Toojinda T, Srinives P (2012) SSR map construction and quantitative trait loci (QTL) identification of major agronomic traits in mungbean (Vigna radiata (L.) Wilczek). SABRAO J Breed Genet 44:71–86
Kasettranan W, Somta P, Srinives P (2010) Mapping of quantitative trait loci controlling powdery mildew resistance in mungbean (Vigna radiate (L.) Wilczek). J Crop Sci Biotech 13:155–161
Kim MY, Kang YJ, Lee T, Lee SH (2013) Divergence of flowering-related genes in three legume species. Plant Gen. doi:10.3835/plantgenome2013.03.0008
Kong F, Liu B, Xia Z, Sato S, Kim BM, Watanabe S, Yamada T, Tabata S, Kanazawa A, Harada K, Abe J (2010) Two coordinately regulated homologs of FLOWERING LOCUS T are involved in the control of photoperiodic flowering in soybean. Plant Physiol 154:1220–1231
Lai J, Li R, Xu X, Jin W, Xu M, Zhao H, Xiang Z, Song W et al (2010) Genome-wide patterns of genetic variation among elite maize inbred lines. Nat Genet 42:1027–1030
Lam HM, Xu X, Liu X, Chen WB, Yang GH, Wong FL, Li MW, He WM, Qin N, Wang B et al (2010) Resequencing of 31 wild and cultivated soybean genomes identifies patterns of genetic diversity and selection. Net Genet 42:1053–1059
Lambrides CJ, Godwin I (2007) Genome mapping and molecular breeding in plants. In: Kole C (ed) Pulses, sugar and tuber crops, vol 3. Springer, Berlin and Heidelberg, pp 69–90
Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10:R25
Liu B, Kanazawa A, Matsumura H, Takahashi R, Harada K, Abe J (2008) Genetic redundancy in soybean photoresponses associated with duplication of the phytochrome A gene. Genetics 180:995–1007
Marcolino-Gomes J, Rodrigues FA, Fuganti-Pagliarini R, Bendix C, Nakayama TJ, Celaya B, Molinari HB, de Oliveira MC, Harmon FG, Nepomuceno A (2014) Diurnal Oscillations of Soybean Circadian Clock and Drought Responsive Genes. PLoS One 9:e86402
Mondal MMA, Fakir MSA, Juraimi AS, Hakim MA, Islam MM, Shamsuddoha ATM (2011) Effects of flowering behavior and pod maturity synchrony on yield of mungbean [Vigna radiata (L.) Wilczek]. Australian J Crop Sci 5:945–953
Moore RC, Purugganan MD (2005) The evolutionary dynamics of plant duplicate genes. Curr Opin Plant Biol 8:122–128
Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5:621–628
Nowrousian M (2010) Next-generation sequencing techniques for eukaryotic microorganisms: sequencing-based solutions to biological problems. Eukaryot Cell 9:1300–1310
Rahman MM (1991) Factors limiting the expansion of summer pulses. In: Sahni BB, Raman U (ed) Advances in pulse research in Bangladesh, Proceedings of the second national workshop on pulses, Gazipur
Smartt J (1990) Grain legume: Evolution and genetic resources: The old world pulses: Vigna species. Cambridge University Press, Cambridge
Tanksley SD, McCouch SR (1997) Seed banks and molecular maps: unlocking genetic potential from the wild. Science 277:1063–1066
Trapnell C, Pachter L, Salzberg SL (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25:1105–1111
Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28:511–515
Wang Y, Wang X, Paterson AH (2012) Genome and gene duplications and gene expression divergence: a view from plants. Ann N Y Acad Sci 1256:1–14
Watanabe S, Hideshima R, Xia Z, Tsubokura Y, Sato S, Nakamoto Y, Yamanaka N, Takahashi R, Ishimoto M, Anai T, Tabata S, Harada K (2009) Map-based cloning of the gene associated with the soybean maturity locus E3. Genetics 182:1251–1262
Watanabe S, Xia Z, Hideshima R, Tsubokura Y, Sato S, Yamanaka N, Takahashi R, Anai T, Tabata S, Kitamura K, Harada K (2011) A map-based cloning strategy employing a residual heterozygous line reveals that the GIGANTEA gene is involved in soybean maturity and flowering. Genetics 188:395–407
Xia Z, Zhai H, Liu B, Kong F, Yuan X, Wu H, Cober ER, Harada K (2012) Molecular identification of genes cotrolling flowering time, maturity, and photoperiod response in soybean. Plant Syst Evol 298:1217–1227
Acknowledgments
This work was carried out with the support of “Cooperative Research Program for Agriculture Science & Technology Development (Project title. PJ008060, Project No. PJ00806003)” Rural Development Administration, Republic of Korea.
Conflict of interest
None declared.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kim, S.K., Lee, T., Kang, Y.J. et al. Genome-wide comparative analysis of flowering genes between Arabidopsis and mungbean. Genes Genom 36, 799–808 (2014). https://doi.org/10.1007/s13258-014-0215-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13258-014-0215-8