Abstract
Circular RNA (circRNA) is a novel class of single-stranded RNAs with a closed loop structure. The majority of circRNAs are formed by a back-splicing process in pre-mRNA splicing. Their expression is dynamically regulated and shows spatiotemporal patterns among cell types, tissues and developmental stages. CircRNAs have important biological functions in many physiological processes, and their aberrant expression is implicated in many human diseases. Due to their high stability, circRNAs are becoming promising biomarkers in many human diseases, such as cardiovascular diseases, autoimmune diseases and human cancers. In this review, we focus on the translational potential of using human blood circRNAs as liquid biopsy biomarkers for human diseases. We highlight their abundant expression, essential biological functions and significant correlations to human diseases in various components of peripheral blood, including whole blood, blood cells and extracellular vesicles. In addition, we summarize the current knowledge of blood circRNA biomarkers for disease diagnosis or prognosis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
INTRODUCTION
Liquid biopsy is a biopsy that uses body liquids as the sample source to diagnose, predict the outcome of or monitor the development of human diseases (Rubis et al., 2019; Luo et al., 2020a). Compared to traditional tissue biopsy, liquid biopsy has the advantages of being noninvasive, performed in real-time and accurate (Rubis et al., 2019; Luo et al., 2020a). It has been proven to be applicable to the management of many human diseases, including cancers (Heitzer et al., 2019; Mattox et al., 2019; Rubis et al., 2019), prenatal genetic disorders (Zhang et al., 2019a), heart diseases (Zemmour et al., 2018), schizophrenia (Chen et al., 2020b), transplant rejection (Bloom et al., 2017) and infectious diseases (Burnham et al., 2018; Hong et al., 2018; Blauwkamp et al., 2019; Han et al., 2020a). To date, most liquid biopsy studies have focused on its clinical application in human cancers (reviewed in Siravegna et al., 2017; Heitzer et al., 2019; Mattox et al., 2019; Rubis et al., 2019). For example, a cell-free DNA (cfDNA)-based liquid biopsy test that determines the mutational status of the epidermal growth factor receptor (EGFR) gene was used to guide the response of EGFR tyrosine kinase inhibitors in non-small cell lung cancer (NSCLC) patients, which was approved by the FDA in clinical practice (Kwapisz, 2017). Another FDA-approved liquid biopsy test, Epi proColon, assessed the methylation status of the Septin9 gene in whole blood, which was used to screen colorectal cancer patients from healthy controls (Lamb and Dhillon, 2017). In addition to cfDNA (Wan et al., 2017; Cescon et al., 2020), several other analytes within circulating body fluids were investigated as liquid biopsy biomarkers, such as circulating tumor cells (CTCs) (Yu et al., 2013), extracellular vesicles (EVs) (Torrano et al., 2016), cell-free RNA (cfRNA) (Zaporozhchenko et al., 2018), circulating proteins (Surinova et al., 2015), circulating metabolites (Crutchfield et al., 2016) and platelets (Joosse and Pantel, 2015; Best et al., 2017). Among them, RNA-based liquid biopsy biomarkers have gained much more attention in recent years since they have dynamic expressions and are closely related to different disease conditions (Zaporozhchenko et al., 2018; Sole et al., 2019). Circulating noncoding RNAs, especially microRNAs (miRNAs), have shown promising potential as stable blood-based biomarkers in liquid biopsy (Mitchell et al., 2008; Anfossi et al., 2018; Pardini et al., 2019).
Circular RNAs (circRNAs) are a group of endogenous noncoding RNA molecules (Chen, 2016, 2020; Li et al., 2018d). They were first found in plant viroids (Sanger et al., 1976) and eukaryotic cells (Hsu and Coca-Prados, 1979) in 1970s, and were recently observed to be functional (Hansen et al., 2013; Memczak et al., 2013). They are joined head to tail to generate a covalently closed loop structure through back-splicing (Vicens and Westhof, 2014; Li et al., 2018d). They lack a 5-prime cap and 3-prime poly-A tail, which is quite different from canonical linear RNAs (Vicens and Westhof, 2014; Li et al., 2018d). CircRNAs have been identified in almost all organisms across the eukaryotic tree of life (Wang et al., 2014). Some circRNAs are the predominant transcript isoform of their host genes expressed in specific tissues or cell types (Salzman et al., 2012, 2013; Rybak-Wolf et al., 2015). Furthermore, they are expressed in a tissue- (Zhou et al., 2018a), cell-type- (Salzman et al., 2013) and developmental stage-specific manner (Zhou et al., 2018a). Accumulating studies have revealed many regulatory roles and versatile cellular functions that circRNAs can perform (Li et al., 2018d; Chen, 2020). They can act as miRNA decoys (Hansen et al., 2013), RNA binding protein (RBP) sponges (Ashwal-Fluss et al., 2014; Huang et al., 2020a) and protein scaffolds (Li et al., 2015c, 2019a). Moreover, a small percentage of circRNAs can be translated into proteins (Legnini et al., 2017; Pamudurti et al., 2017; Heesch et al., 2019). Aberrant expression of circRNAs has been related to many human diseases, including cancers (Shang et al., 2019; Vo et al., 2019), neurodegenerative diseases, cardiovascular diseases (Aufiero et al., 2019) and immune diseases (Chen et al., 2019c; Zhou et al., 2019b). Due to their high stability (Enuka et al., 2015), abundant expression (Jeck et al., 2013) and high specificity (Zhou et al., 2018a), circRNAs are becoming promising biomarkers for human diseases (Zhang et al., 2018j).
Due to the importance of liquid biopsy biomarkers in precision medicine (Vargas and Harris, 2016) and the superior characteristics of circRNAs as disease biomarkers (Zhang et al., 2018j), recent studies have put great efforts into researching the use of circRNAs as liquid biopsy biomarkers for human diseases. Here, we review current progress that has been made on this topic. We focus on circRNAs in the peripheral blood, although circRNAs are abundantly expressed in other body fluids, such as saliva (Bahn et al., 2014; Ghods, 2018) and urine (Kölling et al., 2019; Lam and Lo, 2019; Vo et al., 2019). In this review, we first briefly introduce the biogenesis, function, and expression patterns of circRNAs and their close correlations to human diseases. Then, we emphasize the functional roles they may play in different blood components, including blood cells, serum, plasma, platelets and EVs in the blood. We summarize peripheral blood circRNA biomarkers that have been constructed for the management of human diseases. Finally, we discuss future opportunities and challenges of translating blood circRNAs into clinical practice.
THE BIOGENESIS OF CIRCRNAS
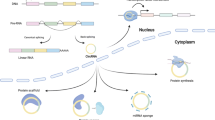
CircRNAs are derived from pre-mRNAs and formed by back-splicing, in which a downstream 5-prime splice site (ss) is covalently joined with an upstream 3-prime ss (Fig. 1) (Li et al., 2018d). There are three major types of circRNAs, exonic circRNAs (ecircRNAs), exon-intron circRNAs (EIcircRNAs) and circular intronic RNAs (ciRNAs) (Fig. 1). EcircRNAs only contain exons, and most ecircRNAs are located in the cytoplasm. Two models have been proposed to explain the production of ecircRNAs (Fig. 1A) (Chen, 2016; Wu et al., 2017a; Kristensen et al., 2019; Xiao et al., 2020). The first model suggests that when the pre-mRNA is partially folded during transcription, canonical splicing will cause an “exon jump” and generate a linear RNA containing skipped exons (Starke et al., 2014; Kelly et al., 2015). A subsequent back-splicing event will turn the ‘lariat intermediate’ into a closed circular transcript (Barrett et al., 2015; Li et al., 2018d). The second model proposes that when the 5-prime ss is pulled closer to the 3-prime ss by base pairing of flanking intronic complementary sequences (Ivanov et al., 2014; Vicens and Westhof, 2014; Zhang et al., 2014; Wilusz, 2015) or specific bindings of intronic sequences to RBPs (Conn et al., 2015), a back-splicing event may occur, and a circRNA may be generated (Jeck et al., 2013). These models are supported by accumulating evidence showing that cis-acting elements (Jeck et al., 2013; Zhang et al., 2014; Yoshimoto et al., 2019) and trans-acting factors (Ashwal-Fluss et al., 2014; Conn et al., 2015; Agirre et al., 2019) play crucial regulatory roles in circRNA formation. EIcircRNAs are generated when an intron or several introns are alternatively retained in splicing (Li et al., 2015c). Therefore, EIcircRNAs are mainly considered intermediates of ecircRNAs (Li et al., 2015c). CiRNAs, which only contain introns, are produced through escape from debranching of intron lariats (Zhang et al., 2013). This process depends on a GU-rich motif near the 5-prime ss and a C-rich motif close to the branch-point site (Zhang et al., 2013). Unlike ecircRNAs, EIcircRNAs and ciRNAs are mainly located in the nucleus.
The biogenesis and function of circRNAs. (A) CircRNAs are formed by back-splicing of pre-mRNAs by different mechanisms, including: lariat driven circularization, intron pairing driven circulation, RBP driven circularization, and debranching escape of intron lariats. (B) CircRNAs can perform diverse biological functions. First, EIcircRNAs can interact with RNA Pol II and U1 snRNP to regulate gene transcription in the nucleus (i). Second, ecircRNAs can accumulate in the cytoplasm and act as miRNA decoys (ii), protein regulators (iii), and translation templates (iv). Third, circRNAs can be secreted into EVs by many cell types and transported to recipient cells by EVs. EV circRNAs can also act as important gene regulators
THE FUNCTION OF CIRCRNAS
With the rapid application of high-throughput RNA-sequencing (RNA-seq) technology (Stark et al., 2019) and related bioinformatics tools that identify circRNAs from RNA-seq datasets (Jeck and Sharpless, 2014; Szabo and Salzman, 2016; Li et al., 2017a; Gao and Zhao, 2018), more than one million circRNAs have been annotated in model organisms (Glažar et al., 2014; Vo et al., 2019; Cai et al., 2020; Wu et al., 2020a). However, the functional characterization of circRNAs is still at its early stage (Chen, 2016, 2020; Li et al., 2018d). Their circular conformation and the large sequence overlap with their linear mRNA counterparts have posed substantial challenges in investigating the functions of circRNAs (Li et al., 2018d). Based on our current knowledge, circRNAs have diverse functions as miRNA decoys, protein regulators and translation templates (Fig. 1B, see (Chen, 2016, 2020; Li et al., 2018d) for excellent reviews).
MiRNA decoys
The most well-known function of circRNAs is that they can act as a sponge to inhibit the function of miRNAs and indirectly regulate the expression of miRNA target genes at the posttranscriptional level (Fig. 1B) (Hansen et al., 2013; Memczak et al., 2013; Weng et al., 2017). For example, Cdr1as can compete with mRNAs by binding miR-7 through miRNA response elements (MREs) (Hansen et al., 2013). A follow-up in vivo experiment that knocked down the Cdr1as locus in the mouse genome confirmed the importance of the Cdr1as and miR-7 interaction in normal brain function (Piwecka et al., 2017). Although it may not be a common phenomenon for most circRNAs (Guo et al., 2014), several abundant circRNAs also function as miRNA sponges, including circASAP1 (Hu et al., 2019c), circBIRC6 (Yu et al., 2017), circHIPK2 (Huang et al., 2017b), circHIPK3 (Zheng et al., 2016) and circSry (Hansen et al., 2013).
Protein regulators
In addition to miRNAs, circRNAs can interact with proteins or protein complexes to regulate protein expression and function (Fig. 1B) (Huang et al., 2020a). First, circRNAs can bind to some RBPs and act as RBP sponges. CircMbl, a circRNA originating from the second exon of the splicing factor muscleblind (MBL), is the first circRNA that was discovered to function as an RBP sponge (Ashwal-Fluss et al., 2014). It was observed that both the circular exon and the flanking introns of circMbl contain many MBL binding sites that can be specifically bound by MBL (Ashwal-Fluss et al., 2014). Therefore, increased expression of MBL can promote back-splicing of circMbl and lead to decreased expression of canonical linear MBL transcripts. At the same time, circMbl can sequester MBL in the cytoplasm and prevent it from performing splicing functions (Ashwal-Fluss et al., 2014). Several other circRNAs, such as circAmotl1 (Yang et al., 2017b), circANRIL (Holdt et al., 2016), circPABPN1 (Abdelmohsen et al., 2017), circPOLR2A (Li et al., 2017e) and circDHX34 (Li et al., 2017e), can function as protein decoys as well. Second, circRNAs can form a complex with other RNAs and proteins to perform functions. For example, some intron-containing circRNAs, such as circEIF3J and circBPTF, can interact with U1 snRNP to form an RNA-protein complex, which further interacts with RNA polymerase II (Pol II) at the promoter region of their parental genes and enhances their transcription (Zhang et al., 2013; Li et al., 2015c). Similarly, some ciRNAs, such as ci-ankrd52, can also associate with Pol II as well (Zhang et al., 2013; Li et al., 2015c). They can also regulate the expression of their parental gene by modulating the elongation of Pol II (Zhang et al., 2013; Li et al., 2015c). Many circRNAs, rather than a specific circRNA, can interact with NF90/NF110 as a group to form circRNP complexes in the cytoplasm, which may function as a reservoir of NF90/NF110 (Li et al., 2017e). Upon viral invasion, NF90/NF110 could be released from circRNP complexes and bind to viral mRNAs to perform its antiviral functions (Li et al., 2017e). Third, the interaction of circRNAs with proteins can facilitate or block the function of their binding proteins. For instance, increased expression of circFoxo3 in the cytoplasm may arrest the anti-senescent protein ID1, transcription factor E2F1, anti-stress proteins FAK and HIF1α and prevent the nuclear translocation of these transcription factors and their function, thus leading to increased cellular senescence (Du et al., 2017). CircFoxo3 can also form a tertiary circRNA-protein complex with cyclin-dependent kinase 2 (CDK2) and cyclin-dependent kinase inhibitor 1 (p21), which facilitates the inhibition of CDK2 by p21, thus blocking cell cycle progression (Du et al., 2016a). Moreover, circFoxo3 can bind to Mdm2 and p53 in a breast cancer cell line, which leads to tumor cell apoptosis (Du et al., 2016b).
Translation
Some circRNAs with an internal ribosome entry site (IRES), such as circMbl (Pamudurti et al., 2017) and circZNF609 (Legnini et al., 2017), can share the start codon of their host genes and be translated in a cap-independent manner. The translation initiation of circRNAs may be promoted by N6-methyladenosine (m6A) modification of circRNAs (Yang et al., 2017c). In the human heart, 40 circRNAs were observed to be associated with ribosomes and thus may be translated (Heesch et al., 2019). Some of them, including circSLC8A1, circMYBPC3 and circRYR2, are heart-specific circRNAs (Heesch et al., 2019). The translation of these circRNAs may be related to the physiological roles of the human heart. Further functional analysis observed that some circRNA-encoded proteins can exert agonist or antagonist effects on cancer progression. For example, PINT87aa (Zhang et al., 2018d) and AKT3-A74aa (Xia et al., 2019), two peptides encoded by circRNAs, can interact with signal factors and inhibit glioblastoma tumorigenicity in glioblastoma. However, circGprc5a-secreted peptides can promote bladder oncogenesis and metastasis through its binding to Gprc5a (Gu et al., 2018).
CIRCRNA EXPRESSION AND ITS IMPLICATION IN DISEASES
Given the important functions that circRNAs can perform, their expression may provide significant clues in understanding the underlying mechanisms of biological processes and disease states. Previous studies have observed that circRNAs are widely expressed in all tissues (Zhou et al., 2018a; Ji et al., 2019; Wu et al., 2020a) and cell types (Salzman et al., 2013) of nearly all species across the eukaryotic tree of life (Wang et al., 2014). Specifically, circRNAs are enriched in brain samples (Westholm et al., 2014; Rybak-Wolf et al., 2015; Szabo et al., 2015; Venø et al., 2015) and human blood samples (Memczak et al., 2015), including peripheral blood mononuclear cells (PBMCs) (Qian et al., 2018), erythrocytes (Alhasan et al., 2016), platelets (Alhasan et al., 2016) and exosomes (Li et al., 2015b). Furthermore, circRNAs are expressed in tissue- (Guo et al., 2014; Szabo et al., 2015; Zhou et al., 2018a) or cell-type- (Salzman et al., 2013) specific and age-dependent manner (Rybak-Wolf et al., 2015; Szabo et al., 2015; You et al., 2015; Zhou et al., 2018a). Using the rat BodyMap dataset, we observed tissue-specific circRNA expression in all 11 rat tissues, which may be closely related to the physiological functions of those tissues (Zhou et al., 2018a). The dynamic expression of circRNAs across time has also been correlated with neuronal differentiation (Rybak-Wolf et al., 2015), neural development (Szabo et al., 2015; You et al., 2015), human terminal B cell differentiation (Agirre et al., 2019) and spermatogenesis (Zhou et al., 2018a).
The spatiotemporal expression of circRNAs is mediated by the balance between circRNA generation and circRNA turnover, which can be regulated at both the transcriptional and posttranscriptional levels (Li et al., 2018d; Chen, 2020). On the one hand, the production of circRNAs can be regulated by intronic complement sequences (ICSs) (Jeck et al., 2013; Liang and Wilusz, 2014; Zhang et al., 2014), cis-splicing elements (Ashwal-Fluss et al., 2014; Starke et al., 2014; Wang and Wang, 2015) and trans-acting splicing factors (Ashwal-Fluss et al., 2014; Conn et al., 2015; Kramer et al., 2015; Khan et al., 2016; Aktaş et al., 2017; Errichelli et al., 2017; Fei et al., 2017; Li et al., 2017e). On the other hand, the degradation of circRNAs can be affiliated by miRNA-initiated AGO2 cleavage (Kleaveland et al., 2018) and nuclease-mediated cleavage, including RNase L endonuclease (Liu et al., 2019a), RNase P/MRP endonuclease complex (Park et al., 2019) and G3BP1 endonuclease (Fischer et al., 2020). Dysregulation of circRNA generation or turnover may lead to aberrant circRNA expression in cells or tissues, which may be related to many human diseases (Chen et al., 2016). Cdr1as, one of the most studied circRNAs, harbors more than 70 binding sites for miR-7, and its overexpression strongly inhibits the activity of the tumor suppressor miR-7 (Hansen et al., 2013). Accumulating evidence has demonstrated its significant increase in colorectal cancer samples, which can inhibit miR-7 function and activate EGFR and RAF1 oncogenes (Weng et al., 2017). Moreover, the dysregulation of the miR-7/Cdr1as axis is involved in some other diseases, such as Alzheimer’s diseases (Lukiw, 2013), diabetes (Xu et al., 2015) and cardiometabolic diseases (Geng et al., 2016). Furthermore, the loss of the Cdr1as locus causes miRNA deregulation and affects synaptic transmission in knockout mice (Piwecka et al., 2017). In addition to Cdr1as, other circRNAs are also implicated in many different human diseases, including cancers (Zhang et al., 2018j; Chen et al., 2019b; Smid et al., 2019; Su et al., 2019; Vo et al., 2019), cardiovascular diseases (Zhang et al., 2018j; Aufiero et al., 2019), neurodevelopment and neurodegenerative diseases (Kumar et al., 2017; Zhang et al., 2018j; Dube et al., 2019; Chen et al., 2020d; Hanan et al., 2020) and immune diseases (Chen et al., 2019c; Zhou et al., 2019b).
CIRCRNAS ARE PROMISING BIOMARKERS FOR HUMAN DISEASES
In general, biomarkers are physiological indexes or biological molecules that can be objectively measured and can indicate either a normal or a pathogenic state (Lesko and Atkinson, 2001). Technological advances in genomics, transcriptomics, proteomics and metabolomics have led to the discovery and validation of many biomarkers that are pushing personalized medicine forward (Chen et al., 2012; Karczewski and Snyder, 2018; Ahadi et al., 2020). A good biomarker with clinical significance must meet the criteria of analytical validity, clinical validity and clinical utility (Byron et al., 2016). As a novel type of noncoding RNA, circRNA has several distinct advantages over canonical linear RNAs as a disease biomarker (Zhang et al., 2018j). First, circRNA is more stable than linear RNAs because it has a closed loop structure without 5-prime and 3-prime ends (Enuka et al., 2015; Li et al., 2015b). In the MCF10A human mammary epithelial cell line, Enuka et al. observed at least 2.5 times longer half-lives of circRNAs compared to linear RNAs, including mRNAs and miRNAs (Enuka et al., 2015). Second, circRNAs are abundantly expressed in many tissue samples. For example, brain samples express more than 10,000 circRNAs in the rat BodyMap dataset (Zhou et al., 2018a). In some cases, the expression values of circRNAs are much higher than their linear counterparts (Westholm et al., 2014; Rybak-Wolf et al., 2015; You et al., 2015; Liang et al., 2017). Third, circRNAs are expressed in a tissue-specific (Salzman et al., 2013; Guo et al., 2014; Szabo et al., 2015; Zhou et al., 2018a) and developmental stage-specific manner (Rybak-Wolf et al., 2015; Szabo et al., 2015; You et al., 2015; Zhou et al., 2018a). Importantly, the tissue specificity of circRNA is higher than that of mRNA of its host gene (Guo et al., 2014; Zhou et al., 2018a). These features suggest that circRNAs may have better analytical validity (Zhang et al., 2018j), including analytical specificity, robustness, reproducibility and repeatability (Byron et al., 2016), when used as biomarker molecules. In a recent study, Maass et al. investigated circRNA expression in 20 clinically relevant tissue samples, which underscored the feasibility of using circRNAs as potential disease biomarkers (Maass et al., 2017).
To date, many circRNAs have been identified as biomarkers for human diseases (Zhang et al., 2018j), especially human cancers (Li et al., 2015a; Meng et al., 2017; Su et al., 2019; Sheng et al., 2020). For example, Cdr1as has been revealed as a prognostic biomarker in colorectal cancer patients (Weng et al., 2017). In a training cohort comprising 153 primary colorectal cancer tissues and 44 matched normal mucosae, significantly increased Cdr1as expression was observed in colorectal cancer tissues, and its overexpression was associated with poor patient survival. The prognostic power of Cdr1as was further validated in an independent validation cohort comprising 165 colorectal cancer patients (Weng et al., 2017). A four-circRNA signature, consisting of hsa_circ_101308, hsa_circ_104423, hsa_circ_104916 and hsa_circ_100269, has been constructed that can predict the early recurrence of stage III gastric cancer (GC) patients after surgery (Zhang et al., 2017). Moreover, Vo et al. constructed a comprehensive catalog of circRNAs in human cancer tissues in a systematic analysis of more than 2,000 cancer samples (Vo et al., 2019). They identified two circRNAs, circ-AURKA and circ-AMACR, as potential diagnostic biomarkers for neuroendocrine prostate cancer (Vo et al., 2019). Although these circRNA biomarkers are potentially useful in cancer management, most of them are derived from tissue samples (Li et al., 2015a; Meng et al., 2017; Su et al., 2019; Sheng et al., 2020). To improve biomarker accessibility, especially for biomarkers of cancer screening and diagnosis, circRNA biomarkers in body fluids are ideal for clinical application (Anfossi et al., 2018). Hence, the authors further investigated and validated the reliability of detecting circRNA biomarkers in urine samples of prostate cancer patients (Vo et al., 2019), which suggests that circRNA in urine samples is a promising strategy for prostate cancer screening.
BLOOD CIRCRNAS AND LIQUID BIPOSY BIOMARKERS
Liquid biopsy has been a revolutionary tool in disease management, supporting the diagnosis, prognosis and treatment guidance of human diseases (Heitzer et al., 2019; Mattox et al., 2019; Rubis et al., 2019; Luo et al., 2020a). In comparison to urine, saliva or cerebrospinal fluid, peripheral blood has been used as the major body fluid in liquid biopsy (Rubis et al., 2019; Luo et al., 2020a). In the circulating blood, aberrantly expressed RNAs or fusion transcripts in different blood components have been associated with human cancers (Byron et al., 2016; Zaporozhchenko et al., 2018; Sole et al., 2019) and infectious diseases (Byron et al., 2016; Correia et al., 2017). These RNA biomarkers include cfRNAs in plasma or serum (Mitchell et al., 2008; Fehlmann et al., 2020), exosome-derived RNAs (Maas et al., 2017), EV-incorporated cfRNAs (Maas et al., 2017) and RNA transcripts in tumor-educated platelets (Best et al., 2017, 2018). These cell-free or EV-incorporated RNA biomarkers represent the changes in expression that occur in abnormal cells, such as dysregulated genes in cancer cells (Byron et al., 2016; Rubis et al., 2019; Luo et al., 2020a). Other than cfRNAs, gene expression profiles of PBMCs or whole blood have been proven to be good indicators of many human diseases (Chaussabel and Baldwin, 2014; Chaussabel, 2015), since they can assess the immune status (Chaussabel, 2015). Unlike the expression of cfRNAs in serum, plasma or EVs, RNA expression levels in PBMCs or whole blood are measures of the host response to exogenous pathogens or autoantigens (Schnell et al., 2018; Shaked, 2019). Several whole blood or PBMC gene expression signatures have been developed for cancer management, including the early diagnosis of colorectal cancer (Marshall et al., 2010; Ciarloni et al., 2015, 2016) and lung cancer (Showe et al., 2009; Kossenkov et al., 2018), the prognosis of adult acute myeloid leukemia (Bullinger et al., 2004; Valk et al., 2004) and prostate cancer patients (Ross et al., 2012), and the monitoring of renal cell carcinoma relapse (Giraldo et al., 2017). Moreover, whole blood or PBMC RNA signatures have been widely investigated as liquid biopsy diagnostic tools for infectious diseases (Ramilo and Mejias, 2017), such as discriminating influenza from other respiratory viral infections (Zaas et al., 2009), differentiating viral and bacterial infections (Tsalik et al., 2016), diagnosing septic patients (Sweeney et al., 2018; Gunsolus et al., 2019; Mayhew et al., 2020), and discriminating active tuberculosis (TB) patients from patients with latent TB or other lung diseases (Bloom et al., 2013; Anderson et al., 2014; Qian et al., 2016; Zak et al., 2016; Sambarey et al., 2017; MacLean et al., 2019; Warsinske et al., 2019; Esmail et al., 2020; Gupta et al., 2020). In addition, the transcriptome of blood cells has been implicated in assessing the likelihood of developing obstructive coronary artery disease in symptomatic nondiabetic patients (Vargas et al., 2013) and predicting antibody-mediated kidney allograft rejection in kidney transplant patients (Loon et al., 2019).
With regard to RNA molecules, both protein-coding mRNAs (Byron et al., 2016; Sole et al., 2019) and several classes of noncoding RNAs (Byron et al., 2016; Anfossi et al., 2018; Pardini et al., 2019; Sole et al., 2019) have been used as blood disease biomarkers. In comparison to mRNAs or long noncoding RNAs, small noncoding RNAs, such as miRNAs (Mitchell et al., 2008; Max et al., 2018; Fehlmann et al., 2020) and noncanonical small RNAs (Fritz et al., 2016; Pardini et al., 2019), have the advantage of high stability in the circulating blood (Mitchell et al., 2008; Anfossi et al., 2018). This superiority is especially important when translating RNA biomarkers into clinical practice (Byron et al., 2016; Anfossi et al., 2018), since fast mRNA degradation in blood sample processing may affect the performance of mRNA biomarkers (Dvinge et al., 2014). Given the high stability of circRNAs, great efforts and substantial progress have been made to investigate the possibility of using circRNA biomarkers in liquid biopsy in recent years.
Next, we summarize circRNAs in the peripheral blood, their correlations with human diseases and their potential application in liquid biopsy as disease biomarkers (Fig. 2). We classified blood circRNAs into blood cell-free circRNAs and circRNAs in blood cells since they have distinct biological meanings in the context of human diseases. Blood cell-free circRNAs, including circulating cell-free circRNAs in plasma, serum and blood EVs, are secreted from different tissue cells into the blood. Therefore, cell-free circRNAs have corresponding tissue origins, and cell-free circRNA biomarkers represent their clinical significance in the original tissue (Li et al., 2019d, 2020). On the other hand, blood cell circRNAs consist of circRNAs in various blood cells, such as monocytes, erythrocytes, neutrophils and platelets. CircRNAs in mixtures of different blood cells, such as circRNAs in lymphocytes, PBMCs and whole blood, are classified in this group as well. The circRNA expression profiles of peripheral blood cells or a mixture of blood cells are important indicators of the host’s immune status (Chaussabel, 2015), which may undergo dynamic changes during acute events such as viral infections (Chen et al., 2018a; Rose et al., 2019; Zhou et al., 2019a). Hence, circRNA biomarkers in blood cells, PBMCs or whole blood represent the specific immune response of an individual to different physiological aspects.
Peripheral blood circRNAs are implicated in human diseases and can be used as potential disease biomarkers in liquid biopsy. (A) Peripheral blood circRNAs are abundantly expressed and can be reliably detected in cell-free circulating blood components (such as exosomes, EVs, plasma, and serum) and blood cells (including PBMCs, macrophages, RBCs, and platelets). (B) Some exosomal circRNAs, such as circ-DB (i), circSHKBP1 (ii), and circUHRF1 (iii), are important regulators in oncogenic pathways, while some circRNAs in exosomes, such as circHIPK3 (iv), play key roles in the release of inflammatory cytokines. (C) Peripheral blood circRNAs, both cell-free circRNAs and intracellular circRNAs in blood cells, have potential clinical applications as liquid biopsy biomarkers in many human diseases, such as the diagnosis, prognosis and treatment guidance of many human diseases, including autoimmune diseases, cancers, cardiovascular diseases, immuno-deficiency diseases, infectious diseases, and neurodegenerative diseases
Cell-free circRNAs in peripheral blood
Mounting evidence has demonstrated abundant circRNA expression in blood plasma (Maass et al., 2017; Yi et al., 2018; Smid et al., 2019) and serum (Gu et al., 2017; Maass et al., 2017; Sonnenschein et al., 2019; Sun et al., 2020b) (Fig. 2A). In the circulating blood, EVs, including exosomes and microvesicles, are important blood components that have great diagnostic and prognostic value (Revenfeld et al., 2014). Li et al. investigated circRNA expression in MHCC-LM3 liver cancer cells and cell-derived exosomes and found at least 2-fold circRNA enrichment in exosomes compared to producer cells (Li et al., 2015b). They further found that the expression level of exosomal circRNAs is largely the same after serum incubation at room temperature for up to 24 h (Li et al., 2015b). Using the extracellular vesicle long RNA (exLR) sequencing method, the same group explored the expression profiles of circRNAs in 352 plasma EV samples (Li et al., 2019d). They found 137,196 circRNA candidates that were expressed in normal plasma EV samples, and the circular to linear ratio was significantly higher in EVs than in PBMCs (Li et al., 2019d). A recent study also isolated platelet-derived EVs and demonstrated the selective release of platelet-specific circRNAs in exosomes and microvesicles (Preußer et al., 2018). Based on these data, a database of blood exosomal circRNAs, exoRBase, has been developed to facilitate the development of circRNA signatures in blood exosomes (Li et al., 2017b). In addition, two previous studies have observed that the majority of circulating cell-free miRNAs were associated with AGO2 protein complexes rather than with vesicles in blood plasma (Arroyo et al., 2011; Turchinovich et al., 2011). Like cell-free miRNAs, blood cell-free circRNAs may bind to several RNA-binding proteins or protein complexes as well (Huang et al., 2020a). Therefore, some cell-free circRNAs may not be associated with blood EVs. In a pilot study of circRNA expression profiles in 348 primary breast cancer tissue samples, Smid et al. found that circCNOT2 is a prognostic biomarker of aromatase inhibitor therapy in advanced breast cancer patients (Smid et al., 2019). To investigate the possibility of using circCNOT2 as a noninvasive biomarker, they further amplified circCNOT2 in plasma samples of four breast cancer patients and found variable circCNOT2 expression in all plasma samples (Smid et al., 2019). All these results suggest that cell-free circRNAs in peripheral blood are stable, enriched and detectable.
Blood EVs can be secreted from cells of many different biological systems and can be circulated to recipient cells through the bloodstream (Revenfeld et al., 2014; Lasda and Parker, 2016). Therefore, blood cell-free circRNAs can perform vital roles in many biological processes, such as cancer cell proliferation, cancer metastasis, drug resistance, hemostasis and inflammation (Fig. 2B) (Wang et al., 2019e; Cui et al., 2020; Li et al., 2020; Shang et al., 2020; Xu et al., 2020b). For example, circ-deubiquitination (circ-DB) in adipose-secreted exosomes was found to regulate deubiquitination via the suppression of miR-34a and the activation of deubiquitination-related USP7 in plasma samples of hepatocellular carcinoma (HCC) patients, which could reduce DNA damage and promote HCC cell growth (Zhang et al., 2018b). Exosomal circSHKBP1 was found to promote the progression of GC by regulating the miR-582-3p/HUR/VEGF pathway and suppressing HSP90 degradation (Xie et al., 2020a). CircUHRF1 in plasma exosomes can inhibit the functions of natural killer cells by upregulating the expression of TIM-3 via miR-449c-5p degradation and drive resistance to anti-PD1 immunotherapy in HCC patients (Zhang et al., 2020a). Moreover, exosomal circHIPK3 can prevent ischemic muscle injury by downregulating miR-421 expression, increasing FOXO3a expression, inhibiting pyroptosis and releasing IL-1β and IL-18 (Yan et al., 2020).
Given the biological functions that blood cell-free circRNAs can perform (Wang et al., 2019e; Cui et al., 2020; Li et al., 2020) and their implications in many human diseases, cell-free circRNAs in the peripheral blood have great potential as liquid biopsy biomarkers of human diseases (Fig. 2C) (Anfossi et al., 2018; Zaporozhchenko et al., 2018; Zhang et al., 2018j). To date, many blood cell-free circRNAs have been introduced for cancer management, including early cancer diagnosis, cancer prognosis and prediction of cancer treatment (Preußer et al., 2018; Aufiero et al., 2019; Fraipont et al., 2019; Lu et al., 2019c; Pardini et al., 2019; Sole et al., 2019; Su et al., 2019; Wang et al., 2019e; Beltrán-García et al., 2020; Cui et al., 2020; Li et al., 2020). In a recent study, Luo et al. measured the expression levels of two circRNAs in plasma samples of 231 lung cancer patients and 41 healthy controls using reverse transcription droplet digital PCR (RT-ddPCR) (Luo et al., 2020c). They identified hsa_circ_0000190 as a circRNA biomarker in human blood plasma that can predict the survival outcomes of lung cancer patients (Luo et al., 2020c). Furthermore, the increased expression of plasma hsa_circ_0000190 was also correlated with poor response to systemic therapy and immunotherapy in lung cancer patients (Luo et al., 2020c). Similarly, Li et al. investigated the clinical relevance of serum exosomal circFLI1 in lung cancer patients in a cohort of 61 small cell lung cancer (SCLC) patients and 55 normal subjects. They found that serum exosomal circFLI1 levels were significantly higher in SCLC patients, especially in SCLC patients with distant metastasis (Li et al., 2018b). Notably, they observed that SCLC patients with lower exosomal circFLI1 expression levels experienced longer disease remissions, indicating its prognostic power in SCLC. The authors also suggested that serum exosomal circFLI1 may be used as a biomarker that can monitor the clinical response to chemotherapy in SCLC patients (Li et al., 2018b). By analyzing blood plasma samples of 62 GC patients and 25 healthy controls, Tang et al. proposed a novel circulating diagnostic biomarker of GC, plasma exosomal circKIAA124, that was correlated with clinical TNM stage, lymphatic metastasis and overall survival time of GC patients (Tang et al., 2018). In addition to the aberrant expression of blood cell-free circRNAs, the presence of some fusion circRNAs (f-circRNAs) in the peripheral blood has also been used as a liquid biopsy biomarker. Guarnerio et al. found that f-circRNAs could be produced from cancer-associated chromosomal translocations in cancer cells (Fig. 1), and f-circRNAs could promote cellular transformation, cell viability and resistance upon therapy (Guarnerio et al., 2016). In a systematic analysis of f-circRNAs in localized prostate cancer tissues, Chen et al. observed more f-circRNAs in tumors with worse prognosis (Chen et al., 2019b). Due to the lack of recurrence of these f-circRNAs, the authors suggested that f-circRNAs are good biomarker candidates (Chen et al., 2019b). In NSCLC patients, Tan et al. found that F-circEA, an f-circRNA originating from the EML4-ALK fusion gene, was exclusively expressed in the plasma of patients with the EML4-ALK fusion (Tan et al., 2018). Therefore, plasma F-circEA may serve as a liquid biopsy biomarker to diagnose NSCLC patients with EML4-ALK translocation and guide targeted therapy for NSCLC patients in this subgroup (Tan et al., 2018). More cell-free circRNA biomarkers in the blood are summarized in Table 1.
CircRNAs in blood cells and whole blood
CircRNA expression in blood cells and whole blood, a major source of liquid biopsy samples, has been extensively investigated (Fig. 2A). In a pilot study, Memczak et al. detected thousands of circRNAs in peripheral whole blood samples using RNA-seq (Memczak et al., 2015). They found that the expression levels of these blood circRNAs were comparable to those in circRNA-rich cerebellar tissue (Memczak et al., 2015). In separate blood cell populations, circRNAs were observed to be enriched approximately 100-fold in platelets and anucleate erythrocytes relative to nucleated tissues, such as lung, brain and colon (Alhasan et al., 2016). Gaffo et al. investigated circRNA expression in T cells, B cells and monocytes of healthy subjects and found abundant circRNA expression in these mature blood cells (Gaffo et al., 2019). We also explored the expression landscape of circRNAs in PBMCs and found that the expression level of circRNAs in PBMCs, together with that in platelets, red blood cells (RBCs) and whole blood, is high enough to be detected (Qian et al., 2018). All these results suggest that whole blood (Memczak et al., 2015; Qian et al., 2018), PBMCs (Qian et al., 2018), and several blood cells, including neutrophils (Maass et al., 2017), T cells (Gaffo et al., 2019), B cells (Gaffo et al., 2019), monocytes (Gaffo et al., 2019), RBCs (Alhasan et al., 2016; Qian et al., 2018) and platelets (Maass et al., 2017; Qian et al., 2018; Gaffo et al., 2019), are reliable clinical samples for circRNA profiling in liquid biopsy.
Accumulating evidence has suggested that circRNAs play crucial roles in the immune response and its regulation (Fig. 3) (Chen et al., 2017b, 2019c; Xu et al., 2018b, 2020a; Yang et al., 2018; Zhou et al., 2019b; Awan et al., 2020). First, circRNAs are important regulators of blood cell biogenesis, differentiation and activation (Chen et al., 2019c; Zhou et al., 2019b; Xu et al., 2020a). In a comprehensive study of circRNA expression in hematopoietic progenitors, differentiated lymphoid and myeloid cells, Nicolet et al. observed a cell-type specific pattern of circRNA expression profiles in blood cells, and the type and number of circRNAs increased upon hematopoietic maturation (Nicolet et al., 2018). Moreover, Holdt et al. found that circANRIL can bind to PES1 to impede the generation of pre-rRNA and ribosomes, resulting in the biogenesis of macrophages by p53 activation (Holdt et al., 2016). Second, circRNAs are actively involved in antiviral immune responses (Fig. 3B) (Cadena and Hur, 2017; Awan et al., 2020). For example, Chen et al. showed that exogenous circRNAs released by viruses can be recognized by the pattern recognition receptor RIG-I of host cells, which activates host innate immunity (Chen et al., 2017a). In their subsequent work, the authors showed that m6A RNA modification of human circRNAs inhibits innate immunity, while unmodified circRNAs and K63-polyubiquitin can activate RIG-I and innate immune response (Chen et al., 2019a). Li et al. found that two immune factors, NF90/NF110, not only promote circRNA production in the nucleus but also bind to mature host circRNAs to form circRNP complexes in the cytoplasm. Upon viral infection, circRNP complexes in the cytoplasm can release NF90/NF110, which binds viral mRNAs to inhibit viral replication (Li et al., 2017e). Moreover, Liu et al. presented that circRNAs can form RNA duplexes and act as inhibitors of innate immunity-related protein kinase (PKR) under normal conditions (Liu et al., 2019a). Upon poly(I:C) treatment or viral infection, RNase L is activated to efficiently degrade circRNAs, and PKR is thus released from circRNA inhibition to initiate the early cellular innate immune response (Liu et al., 2019a). Third, circRNAs are closely associated with the antibacterial immune response as well. Ng et al. characterized circRNAs induced by lipopolysaccharide (LPS) and identified circRasGEF1B as a conserved positive regulator of the LPS response (Ng et al., 2016). Their functional analysis revealed that circRasGEF1B can induce the expression of ICAM-1 in the TLR4/LPS pathway, which activates pathogen recognition and the inflammatory response upon microbial infection (Fig. 3A) (Ng et al., 2016).
CircRNAs are actively involved in host immune responses to exogenous pathogens. (A) CircRasGEF1B, a circRNA induced by LPS, can protect cells from bacterial infection by regulating the expression of ICAM-1 mRNA in the TLR4/LPS pathway. (B) Exogenous circRNAs released by viruses can be recognized by RIG-I, thus activating the host innate immunity to viruses (i). Moreover, NF90/NF110 not only promotes circRNA production in the nucleus but also interacts with mature host circRNAs to form circRNP complexes in the cytoplasm. Upon viral infection, NF90/NF110 can be released from circRNP complexes and bind viral mRNAs to inhibit viral replication (ii). In addition, circRNAs can form RNA duplexes and act as inhibitors of PKR under normal conditions. When a virus invades the cells of its host, RNase L is activated to efficiently degrade circRNAs, and PKR is released and activated to initiate the early cellular innate immune response (iii)
The important functions of circRNAs in blood cells suggest that the dysregulation of circRNA expression in blood cells is likely to contribute to the occurrence and progression of immune-related diseases, including autoimmune diseases, infectious diseases and cardiovascular diseases (Chen et al., 2019c; Gaffo et al., 2019; Zhou et al., 2019b; Xie et al., 2020b; Xu et al., 2020a). For instance, we determined that the expression level of PBMC circRNAs is higher in TB patients than healthy controls, and five immune-related pathways were upregulated upon Mycobacterium tuberculosis infection, including “cytokine-cytokine receptor interactions”, “chemokine signaling pathways”, and “neurotrophic signaling pathways” (Qian et al., 2018). Similarly, Huang et al. identified 13 upregulated and 24 downregulated circRNAs in PBMCs of TB patients (Huang et al., 2018c). Among them, hsa_circRNA_001937 is likely to participate in the inflammatory response by targeting miR-26b and modulating the NF-κB pathway (Huang et al., 2018c). In addition, Zhang et al. investigated the roles that circRNAs play in early human immunodeficiency virus (HIV) infection (EHI), especially in regulating HIV replication (Zhang et al., 2018i). EHI represents a stage where viral replication increases to a peak level and intense antiviral immune response and immune injury occur (Powers et al., 2011; Richey and Halperin, 2013). The authors characterized the expression profiles of circRNAs, mRNAs and miRNAs in PBMCs of EHI patients and constructed a circRNA-associated competing endogenous network in EHI patients. They revealed 67 differentially expressed circRNAs, such as CCNK, CDKN1A and IL-15, that are potentially involved in HIV replication by regulating the expression of genes in the immune response, inflammatory response and defense response to the virus (Zhang et al., 2018i). Regarding autoimmune diseases, Liu et al. detected a global reduction in circRNAs and activation of RNase L in PBMCs of systemic lupus erythematosus (SLE) patients (Liu et al., 2019a). They further found that circPOLR2A overexpression can lead to reduced PKR activation, EIF2α phosphorylation and type I IFN-induced gene suppression. This highlights the link between circRNAs and innate immunity regulation and provides the potential for circRNA manipulation in SLE treatment (Liu et al., 2019a). In addition to the above examples, the abnormal expression of blood circRNAs is related to several other diseases, such as the immune response to Ebola virus (Wang et al., 2017b) and hepatitis C virus (Jost et al., 2018), rheumatoid arthritis (RA) (Yang et al., 2019), type 2 diabetes mellitus (T2DM) (Fang et al., 2018), heart failure (Han et al., 2020b) and adenosine deaminase deficiency (Maass et al., 2017).
With increasing knowledge of blood cell circRNAs and their function, many circRNAs in blood cells or whole blood have been proposed as liquid biopsy biomarkers for human diseases (Fig. 2C) (Aufiero et al., 2019; Beltrán-García et al., 2020; Kumar et al., 2017; Ravnik-Glavač and Glavač, 2020; Sun et al., 2020a). For instance, we developed a PBMC circRNA-based molecular signature that discriminates active TB patients from healthy controls in our previous study (Qian et al., 2018). The classification power of this PBMC circRNA signature was further validated in an independent cohort with an area under the receiver operating characteristic curve (AUC) of 0.946 (Qian et al., 2018). In another study, Huang et al. found that the expression of hsa_circ_001937 in PBMCs was significantly higher in TB patients than in pneumonia, chronic obstructive pulmonary disease (COPD) and lung cancer patients (Huang et al., 2018c). In a cohort consisting of 115 TB, 40 pneumonia, 40 COPD, and 40 lung cancer patients and 90 healthy control subjects, hsa_circ_001937 had good discriminative power with an AUC of 0.873. After anti-TB treatment, the expression level of hsa_circ_001937 was significantly decreased compared to that of healthy controls. These results suggest that PBMC hsa_circ_001937 may be a TB diagnostic biomarker (Huang et al., 2018c). Furthermore, Lei et al. found an upregulation of circ_0000798 expression in PBMCs of HCC patients, which was associated with poor overall survival (Lei et al., 2019). In a cohort of 72 HCC patients and 30 healthy control subjects, circ_0000798 expression could distinguish HCC patients from healthy controls with an AUC of up to 0.703. The authors suggested that PBMC circ_0000798 has potential as a blood biomarker for HCC diagnosis and prognosis (Lei et al., 2019). In addition, Li et al. measured circRNA expression changes between children with SLE and healthy children and investigated the significance of blood circRNAs in SLE diagnosis (Li et al., 2019b). They identified and validated the diagnostic power of two circRNAs in whole blood, hsa_circ_0057762 and hsa_circ_0003090, that can differentiate children with SLE from healthy controls (Li et al., 2019b). Zhao et al. also identified and validated hsa_circ_0054633 in peripheral whole blood as a sensitive and specific diagnostic biomarker for prediabetes and T2DM (Zhao et al., 2017b). In addition to the above examples, the potential of using blood circRNAs as disease biomarkers has been explored for many other human diseases, such as coronary artery disease (Zhao et al., 2017a; Wang et al., 2019a; Liang et al., 2020), community‐acquired pneumonia (Zhao et al., 2019), and schizophrenia (Yao et al., 2019). A list of current proposed potential blood circRNA biomarkers is listed in Table 2.
CONCLUSIONS AND FUTURE PERSPECTIVES
The high stability, abundance and spatiotemporal specific expression of blood circRNAs make them ideal biomarkers for liquid biopsy. In the past several years, many studies have shown that blood circRNAs, both cell-free blood circRNAs and circRNAs in blood cells, have great potential as biomarkers of many human diseases in liquid biopsy. A biomarker with broad clinical application must have demonstrated analytical validity, clinical validity and clinical utility (Byron et al., 2016). Therefore, several issues need to be considered and investigated before peripheral blood circRNA biomarkers can be translated into clinical practice. First, a blood circRNA-based gene test should prove its analytical validity within clinically relevant conditions. Although substantial advances have been made in the past several years (Szabo and Salzman, 2016; Kristensen et al., 2019), the methods to discover and profile circRNAs are far from optimal. Future studies need to test the analytical performance of different circRNA profiling methods in clinical blood samples, such as RNA-seq, circRNA microarray, reverse transcription quantitative PCR (RT-qPCR) and RT-ddPCR (Kristensen et al., 2019). In estimating analytical sensitivity and specificity, reference standards that can be specifically applied in circRNA discovery and profiling are needed (Hardwick et al., 2017). Furthermore, the procedure to discover and validate blood circRNA biomarkers needs to be standardized, including blood collection and preservation, circRNA extraction, library construction, and computational analysis (Byron et al., 2016; Anfossi et al., 2018). With the use of a standardized procedure, future studies need to estimate the technical robustness and reproducibility of the proposed biomarkers within and between laboratories. Second, the blood circRNA biomarkers identified in current studies (Tables 1 and 2) are only preliminary biomarker signatures for human diseases. The designed experiments in most studies are case-control studies of samples with well-defined phenotypes, and the sample size is relatively small. To test their clinical validity, more clinical samples are required to validate their sensitivity and specificity in a larger cohort, especially their performance in discriminating patients with similar clinical phenotypes. Moreover, their performance in diagnosing the disease or predicting the disease outcome also needs to be tested in a prospective cohort in a clinical practice setting. Finally, further studies are needed to test and validate the usefulness of these biomarkers in clinical practice, such as their ability to inform clinical decisions and improve outcomes. Although many challenges and problems need to be solved, the promising potential of translating blood circRNA biomarkers into the clinic brings us new and inspiring options for liquid biopsy.
REFERENCES
Abdelmohsen K, Panda AC, Munk R, Grammatikakis I, Dudekula DB, De S, Kim J, Noh J, Kim K, Martindale JL et al (2017) Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol 14:361–369
Agirre X, Meydan C, Jiang Y, Garate L, Doane AS, Li Z, Verma A, Paiva B, Martín-Subero JI, Elemento O et al (2019) Long non-coding RNAs discriminate the stages and gene regulatory states of human humoral immune response. Nat Commun 10:821
Ahadi S, Zhou W, Rose S, Sailani RM, Contrepois K, Avina M, Ashland M, Brunet A, Snyder M (2020) Personal aging markers and ageotypes revealed by deep longitudinal profiling. Nat Med 26:83–90
Aktaş T, Ilık İ, Maticzka D, Bhardwaj V, Rodrigues C, Mittler G, Manke T, Backofen R, Akhtar A (2017) DHX9 suppresses RNA processing defects originating from the Alu invasion of the human genome. Nature 544:115–119
Alhasan AA, Izuogu OG, Al-Balool HH, Steyn JS, Evans A, Colzani M, Ghevaert C, Mountford JC, Marenah L, Elliott DJ et al (2016) Circular RNA enrichment in platelets is a signature of transcriptome degradation. Blood 127:e1–e11
Anderson ST, Kaforou M, Brent AJ, Wright VJ, Banwell CM, Chagaluka G, Crampin AC, Dockrell HM, French N, Hamilton MS et al (2014) Diagnosis of childhood tuberculosis and host RNA expression in Africa. N Engl J Med 370:1712–1723
Anfossi S, Babayan A, Pantel K, Calin GA (2018) Clinical utility of circulating non-coding RNAs—an update. Nat Rev Clin Oncol 15:541–563
Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, Mitchell PS, Bennett CF, Pogosova-Agadjanyan EL, Stirewalt DL et al (2011) Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci USA 108:5003–5008
Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, Kadener S (2014) CircRNA biogenesis competes with pre-mRNA splicing. Mol Cell 56:55–66
Aufiero S, Reckman YJ, Pinto YM, Creemers EE (2019) Circular RNAs open a new chapter in cardiovascular biology. Nat Rev Cardiol 16:503–514
Awan FM, Yang BB, Naz A, Hanif A, Ikram A, Obaid A, Malik A, Janjua HA, Ali A, Sharif S (2020) The emerging role and significance of circular RNAs in viral infections and antiviral immune responses: possible implication as theranostic agents. RNA Biol. https://doi.org/10.1080/15476286.2020.1790198
Bahn JH, Zhang Q, Li F, Chan TM, Lin X, Kim Y, Wong DTW, Xiao X (2014) The landscape of microRNA, piwi-interacting RNA, and circular RNA in human saliva. Clin Chem 61:221–230
Bao X, He X, Zheng S, Sun J, Luo Y, Tan R, Zhao J, Zhong F, Zhang L (2019) Up-regulation of circular RNA hsa_circ_0037909 promotes essential hypertension. J Clin Lab Anal 33:e22853
Bao X, Zheng S, Mao S, Gu T, Liu S, Sun J, Zhang L (2018) A potential risk factor of essential hypertension in case–control study: circular RNA hsa_circ_0037911. Biochem Biophys Res Commun 498:789–794
Barrett SP, Wang PL, Salzman J (2015) Circular RNA biogenesis can proceed through an exon-containing lariat precursor. eLife 4:e07540
Beltrán-García J, Osca-Verdegal R, Nacher-Sendra E, Pallardó FV, García-Giménez JL (2020) Circular RNAs in sepsis: biogenesis, function, and clinical significance. Cells 9:1544
Best MG, Vancura A, Wurdinger T (2017) Platelet RNA as a circulating biomarker trove for cancer diagnostics. J Thromb Haemost 15:1295–1306
Best MG, Wesseling P, Wurdinger T (2018) Tumor-educated platelets as a noninvasive biomarker source for cancer detection and progression monitoring. Cancer Res 78:3407–3412
Blauwkamp TA, Thair S, Rosen MJ, Blair L, Lindner MS, Vilfan ID, Kawli T, Christians FC, Venkatasubrahmanyam S, Wall GD et al (2019) Analytical and clinical validation of a microbial cell-free DNA sequencing test for infectious disease. Nat Microbiol 4:663–674
Bloom CI, Graham CM, Berry MP, Rozakeas F, Redford PS, Wang Y, Xu Z, Wilkinson KA, Wilkinson RJ, Kendrick Y et al (2013) Transcriptional blood signatures distinguish pulmonary tuberculosis, pulmonary sarcoidosis, pneumonias and lung cancers. PLoS ONE 8:e70630
Bloom RD, Bromberg JS, Poggio ED, Bunnapradist S, Langone AJ, Sood P, Matas AJ, Mehta S, Mannon RB, Sharfuddin A et al (2017) Cell-free DNA and active rejection in kidney allografts. J Am Soc Nephrol 28:2221–2232
Bullinger L, Döhner K, Bair E, Fröhling S, Schlenk RF, Tibshirani R, Döhner H, Pollack JR (2004) Use of gene-expression profiling to identify prognostic subclasses in adult acute myeloid leukemia. N Engl J Med 350:1605–1616
Burnham P, Dadhania D, Heyang M, Chen F, Westblade LF, Suthanthiran M, Lee J, Vlaminck I (2018) Urinary cell-free DNA is a versatile analyte for monitoring infections of the urinary tract. Nat Commun 9:2412
Byron SA, Keuren-Jensen KR, Engelthaler DM, Carpten JD, Craig DW (2016) Translating RNA sequencing into clinical diagnostics: opportunities and challenges. Nat Rev Genet 17:257–271
Cadena C, Hur S (2017) Antiviral immunity and circular RNA: no end in sight. Mol Cell 67:163–164
Cai Z, Fan Y, Zhang Z, Lu C, Zhu Z, Jiang T, Shan T, Peng Y (2020) VirusCircBase: a database of virus circular RNAs. Brief Bioinform. https://doi.org/10.1093/bib/bbaa052
Cescon DW, Bratman SV, Chan SM, Siu LL (2020) Circulating tumor DNA and liquid biopsy in oncology. Nat Cancer 1:276–290
Chaussabel D (2015) Assessment of immune status using blood transcriptomics and potential implications for global health. Semin Immunol 27:58–66
Chaussabel D, Baldwin N (2014) Democratizing systems immunology with modular transcriptional repertoire analyses. Nat Rev Immunol 14:271–280
Chen A, Zhong L, Ju K, Lu T, Lv J, Cao H (2020) Plasmatic circRNA predicting the occurrence of human glioblastoma. Cancer Manag Res 12:2917–2923
Chen GY, Chen R, Ahmad S, Verma R, Kasturi S, Amaya L, Broughton JP, Kim J, Cadena C, Pulendran B et al (2019) N6-methyladenosine modification controls circular RNA immunity. Mol Cell 76:96-109.e109
Chen GY, Kim MV, Chen X, Batista PJ, Aoyama S, Wilusz JE, Iwasaki A, Chang HY (2017) Sensing self and foreign circular RNAs by intron identity. Mol Cell 67:228–238
Chen GY, Satpathy AT, Chang HY (2017) Gene regulation in the immune system by long noncoding RNAs. Nat Immunol 18:962–972
Chen J, Zang Z, Braun U, Schwarz K, Harneit A, Kremer T, Ma R, Schweiger J, Moessnang C, Geiger L et al (2020) Association of a reproducible epigenetic risk profile for schizophrenia with brain methylation and function. JAMA Psychiatry 77(6):628–636
Chen L-L (2016) The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol 17:205–211
Chen L-L (2020) The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat Rev Mol Cell Biol 21:1–16
Chen R, Mias GI, Li-Pook-Than J, Jiang L, Lam H, Chen R, Miriami E, Karczewski KJ, Hariharan M, Dewey FE et al (2012) Personal omics profiling reveals dynamic molecular and medical phenotypes. Cell 148:1293–1307
Chen R, Xia L, Tu K, Duan M, Kukurba K, Li-Pook-Than J, Xie D, Snyder M (2018) Longitudinal personal DNA methylome dynamics in a human with a chronic condition. Nat Med 24:1930–1939
Chen S, Huang V, Xu X, Livingstone J, Soares F, Jeon J, Zeng Y, Hua J, Petricca J, Guo H et al (2019) Widespread and functional RNA circularization in localized prostate cancer. Cell 176:831-843.e822
Chen S, Li T, Zhao Q, Xiao B, Guo J (2017) Using circular RNA hsa_circ_0000190 as a new biomarker in the diagnosis of gastric cancer. Clin Chim Acta 466:167–171
Chen W, Quan Y, Fan S, Wang H, Liang J, Huang L, Chen L, Liu Q, He P, Ye Y (2020) Exosome-transmitted circular RNA hsa_circ_0051443 suppresses hepatocellular carcinoma progression. Cancer Lett 475:119–128
Chen X, Chen RX, Wei WS, Li YH, Feng ZH, Tan L, Chen JW, Yuan GJ, Chen SL, Guo SJ et al (2018) PRMT5 circular RNA promotes metastasis of urothelial carcinoma of the bladder through sponging miR-30c to induce epithelial–mesenchymal transition. Clin Cancer Res 24:6319–6330
Chen X, Yang T, Wang W, Xi W, Zhang T, Li Q, Yang A, Wang T (2019) Circular RNAs in immune responses and immune diseases. Theranostics 9:588–607
Chen Y, Li C, Tan C, Liu X (2016) Circular RNAs: a new frontier in the study of human diseases. J Med Genet 53:359–365
Chen Y-J, Chen C-Y, Mai T-L, Chuang C-F, Chen Y-C, Gupta SK, Yen L, Wang Y-D, Chuang T-J (2020) Genome-wide, integrative analysis of circular RNA dysregulation and the corresponding circular RNA–microRNA–mRNA regulatory axes in autism. Genome Res 30:375–391
Chi BJ, Zhao DM, Liu L, Yin XZ, Wang FF, Bi S, Gui SL, Zhou SB, Qin WB, Wu DM et al (2019) Downregulation of hsa_circ_0000285 serves as a prognostic biomarker for bladder cancer and is involved in cisplatin resistance. Neoplasma 66:197–202
Ciarloni L, Ehrensberger S, Imaizumi N, Monnier-Benoit S, Nichita C, Myung S-J, Kim J, Song S, Kim T, van der Weg B et al (2016) Development and clinical validation of a blood test based on 29-gene expression for early detection of colorectal cancer. Clin Cancer Res 22:4604–4611
Ciarloni, L., Hosseinian, S., Monnier-Benoit, S., Imaizumi, N., Dorta, G., Ruegg, C., and Group, O.b.o.t (2015) Discovery of a 29-gene panel in peripheral blood mononuclear cells for the detection of colorectal cancer and adenomas using high throughput real-time PCR. PLoS ONE 10:e0123904
Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA, Goodall GJ (2015) The RNA binding protein quaking regulates formation of circRNAs. Cell 160:1125–1134
Correia CN, Nalpas NC, McLoughlin KE, Browne JA, Gordon SV, MacHugh DE, Shaughnessy RG (2017) Circulating microRNAs as potential biomarkers of infectious disease. Front Immunol 8:118
Crutchfield CA, Thomas SN, Sokoll LJ, Chan DW (2016) Advances in mass spectrometry-based clinical biomarker discovery. Clin Proteomics 13:1
Cui C, Yang J, Li X, Liu D, Fu L, Wang X (2020) Functions and mechanisms of circular RNAs in cancer radiotherapy and chemotherapy resistance. Mol Cancer 19:58
de Fraipont F, Gazzeri S, Cho WC, Eymin B (2019) Circular RNAs and RNA splice variants as biomarkers for prognosis and therapeutic response in the liquid biopsies of lung cancer patients. Front Genet 10:390
Ding C, Yi X, Wu X, Bu X, Wang D, Wu Z, Zhang G, Gu J, Kang D (2020) Exosome-mediated transfer of circRNA CircNFIX enhances temozolomide resistance in glioma. Cancer Lett 479:1–12
Dolinar A, Koritnik B, Glavac D, Ravnik-Glavac M (2019) Circular RNAs as potential blood biomarkers in amyotrophic lateral sclerosis. Mol Neurobiol 56:8052–8062
Du WW, Fang L, Yang W, Wu N, Awan FM, Yang Z, Yang BB (2016) Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ 24:357–370
Du WW, Yang W, Chen Y, Wu Z-K, Foster FS, Yang Z, Li X, Yang BB (2017) Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur Heart J 38:1402–1412
Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB (2016) Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res 44:2846–2858
Dube U, Del-Aguila JL, Li Z, Budde JP, Jiang S, Hsu S, Ibanez L, Fernandez M, Farias F, Norton J et al (2019) An atlas of cortical circular RNA expression in Alzheimer disease brains demonstrates clinical and pathological associations. Nat Neurosci 22:1–10
Dvinge H, Ries RE, Ilagan JO, Stirewalt DL, Meshinchi S, Bradley RK (2014) Sample processing obscures cancer-specific alterations in leukemic transcriptomes. Proc Natl Acad Sci USA 111:16802–16807
Enuka Y, Lauriola M, Feldman ME, Sas-Chen A, Ulitsky I, Yarden Y (2015) Circular RNAs are long-lived and display only minimal early alterations in response to a growth factor. Nucleic Acids Res 44:1370–1383
Errichelli L, Modigliani S, Laneve P, Colantoni A, Legnini I, Capauto D, Rosa A, Santis R, Scarfò R, Peruzzi G et al (2017) FUS affects circular RNA expression in murine embryonic stem cell-derived motor neurons. Nat Commun 8:14741
Esmail H, Cobelens F, Goletti D (2020) Transcriptional biomarkers for predicting development of tuberculosis: progress and clinical considerations. Eur Respir J 55:1901957
Fan L, Cao Q, Liu J, Zhang J, Li B (2019) Circular RNA profiling and its potential for esophageal squamous cell cancer diagnosis and prognosis. Mol Cancer 18:16
Fang Y, Wang X, Li W, Han J, Jin J, Su F, Zhang J, Huang W, Xiao F, Pan Q et al (2018) Screening of circular RNAs and validation of circANKRD36 associated with inflammation in patients with type 2 diabetes mellitus. Int J Mol Med 42:1865–1874
Fehlmann T, Kahraman M, Ludwig N, Backes C, Galata V, Keller V, Geffers L, Mercaldo N, Hornung D, Weis T et al (2020) Evaluating the use of circulating microRNA profiles for lung cancer detection in symptomatic patients. JAMA Oncol 6(5):714–723
Fei T, Chen Y, Xiao T, Li W, Cato L, Zhang P, Cotter MB, Bowden M, Lis RT, Zhao SG et al (2017) Genome-wide CRISPR screen identifies HNRNPL as a prostate cancer dependency regulating RNA splicing. Proc Natl Acad Sci USA 114:E5207–E5215
Fischer JW, Busa VF, Shao Y, Leung AKL (2020) Structure-mediated RNA decay by UPF1 and G3BP1. Mol Cell 78:70-84.e76
Fritz JV, Heintz-Buschart A, Ghosal A, Wampach L, Etheridge A, Galas D, Wilmes P (2016) Sources and functions of extracellular small RNAs in human circulation. Annu Rev Nutr 36:301–336
Fu Y, Wang J, Qiao J, Yi Z (2019) Signature of circular RNAs in peripheral blood mononuclear cells from patients with active tuberculosis. J Cell Mol Med 23:1917–1925
Gaffo E, Boldrin E, Molin A, Bresolin S, Bonizzato A, Trentin L, Frasson C, Debatin K-M, Meyer LH, te Kronnie et al (2019) Circular RNA differential expression in blood cell populations and exploration of circRNA deregulation in pediatric acute lymphoblastic leukemia. Sci Rep 9:14670
Gao Y, Zhao F (2018) Computational strategies for exploring circular RNAs. Trends Genet. https://doi.org/10.1016/j.tig.2017.12.016
Geng H-H, Li R, Su Y-M, Xiao J, Pan M, Cai X-X, Ji X-P (2016) The circular RNA Cdr1as promotes myocardial infarction by mediating the regulation of miR-7a on its target genes expression. PLoS ONE 11:e0151753
Ghods F (2018) Circular RNA in saliva. Adv Exp Med Biol 1087:131–139
Giraldo NA, Becht E, Vano Y, Petitprez F, Lacroix L, Validire P, Sanchez-Salas R, Ingels A, Oudard S, Moatti A et al (2017) Tumor-infiltrating and peripheral blood T-cell immunophenotypes predict early relapse in localized clear cell renal cell carcinoma. Clin Cancer Res 23:4416–4428
Glažar P, Papavasileiou P, Rajewsky N (2014) circBase: a database for circular RNAs. RNA 20:1666–1670
Gong Y, Mao J, Wu D, Wang X, Li L, Zhu L, Song R (2018) Circ-ZEB1.33 promotes the proliferation of human HCC by sponging miR-200a-3p and upregulating CDK6. Cancer Cell Int 18:116
Gu C, Zhou N, Wang Z, Li G, Kou Y, Yu S, Feng Y, Chen L, Yang J, Tian F (2018) circGprc5a promoted bladder oncogenesis and metastasis through Gprc5a-targeting peptide. Mol Ther Nucleic Acids 13:633–641
Gu Y, Ke G, Wang L, Zhou E, Zhu K, Wei Y (2017) Altered expression profile of circular RNAs in the serum of patients with diabetic retinopathy revealed by microarray. Ophthalmic Res 58:176–184
Guarnerio J, Bezzi M, Jeong J, Paffenholz SV, Berry K, Naldini MM, Lo-Coco F, Tay Y, Beck AH, Pandolfi P (2016) Oncogenic role of fusion-circRNAs derived from cancer-associated chromosomal translocations. Cell 165:289–302
Gunsolus IL, Sweeney TE, Liesenfeld O, Ledeboer NA (2019) Diagnosing and managing sepsis by probing the host response to infection: advances, opportunities, and challenges. J Clin Microbiol 57:e00425-e1419
Guo JU, Agarwal V, Guo H, Bartel DP (2014) Expanded identification and characterization of mammalian circular RNAs. Genome Biol 15:409
Guo G, Wang H, Ye L, Shi X, Yan K, Lin K, Huang Q, Li B, Lin Q, Zhu L et al (2019) Hsa_circ_0000479 as a novel diagnostic biomarker of systemic lupus erythematosus. Front Immunol 10:2281
Gupta RK, Turner CT, Venturini C, Esmail H, Rangaka MX, Copas A, Lipman M, Abubakar I, Noursadeghi M, Gupta RK et al (2020) Concise whole blood transcriptional signatures for incipient tuberculosis: a systematic review and patient-level pooled meta-analysis. Lancet Respir Med 8:395–406
Han D, Li R, Shi J, Tan P, Zhang R, Li J (2020a) Liquid biopsy for infectious diseases: a focus on microbial cell-free DNA sequencing. Theranostics 10:5501–5513
Han J, Zhang L, Hu L, Yu H, Xu F, Yang B, Zhang R, Zhang Y, An Y (2020b) Circular RNA-expression profiling reveals a potential role of hsa_circ_0097435 in heart failure via sponging multiple microRNAs. Front Genet 11:212
Han L, Zhang X, Wang A, Ji Y, Cao X, Qin Q, Yu T, Yin L, Huang H (2020c) A dual-circular RNA signature as a non-invasive diagnostic biomarker for gastric cancer. Front Oncol 10:184
Hanan M, Simchovitz A, Yayon N, Vaknine S, Cohen-Fultheim R, Karmon M, Madrer N, Rohrlich TM, Maman M, Bennett ER et al (2020) A Parkinson’s disease circRNAs resource reveals a link between circSLC8A1 and oxidative stress. EMBO Mol Med 12(9):e11942
Hang D, Zhou J, Qin N, Zhou W, Ma H, Jin G, Hu Z, Dai J, Shen H (2018) A novel plasma circular RNA circFARSA is a potential biomarker for non-small cell lung cancer. Cancer Med 7:2783–2791
Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J (2013) Natural RNA circles function as efficient microRNA sponges. Nature 495:384–388
Hardwick SA, Deveson IW, Mercer TR (2017) Reference standards for next-generation sequencing. Nat Rev Genet 18(8):473–484
He F, Zhong X, Lin Z, Lin J, Qiu M, Li X, Hu Z (2020) Plasma exo-hsa_circRNA_0056616: A potential biomarker for lymph node metastasis in lung adenocarcinoma. J Cancer 11:4037–4046
Heitzer E, Haque IS, Roberts CES, Speicher MR (2019) Current and future perspectives of liquid biopsies in genomics-driven oncology. Nat Rev Genet 20:71–88
Holdt LM, Stahringer A, Sass K, Pichler G, Kulak NA, Wilfert W, Kohlmaier A, Herbst A, Northoff BH, Nicolaou A et al (2016) Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat Commun 7:12429
Hong DK, Blauwkamp TA, Kertesz M, Bercovici S, Truong C, Banaei N (2018) Liquid biopsy for infectious diseases: sequencing of cell-free plasma to detect pathogen DNA in patients with invasive fungal disease. Diagn Microbiol Infect Dis 92(3):210–213
Hsu M-T, Coca-Prados M (1979) Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature 280:339–340
Hu X, Wu D, He X, Zhao H, He Z, Lin J, Wang K, Wang W, Pan Z, Lin H et al (2019) circGSK3beta promotes metastasis in esophageal squamous cell carcinoma by augmenting beta-catenin signaling. Mol Cancer 18:160
Hu Y, Zhu Y, Zhang W, Lang J, Ning L (2019) Utility of plasma circBNC2 as a diagnostic biomarker in epithelial ovarian cancer. Onco Targets Ther 12:9715–9723
Hu ZQ, Zhou SL, Li J, Zhou ZJ, Wang PC, Xin HY, Mao L, Luo CB, Yu SY, Huang XW et al (2019) Circular RNA sequencing identifies circASAP1 as a key regulator in hepatocellular carcinoma metastasis. Hepatology. https://doi.org/10.1002/hep.31068
Huang M, He YR, Liang LC, Huang Q, Zhu ZQ (2017a) Circular RNA hsa_circ_0000745 may serve as a diagnostic marker for gastric cancer. World J Gastroenterol 23:6330–6338
Huang R, Zhang Y, Han B, Bai Y, Zhou R, Gan G, Chao J, Hu G, Yao H (2017b) Circular RNA HIPK2 regulates astrocyte activation via cooperation of autophagy and ER stress by targeting MIR124-2HG. Autophagy 13:1722–1741
Huang Z, Su R, Deng Z, Xu J, Peng Y, Luo Q, Li J (2017c) Identification of differentially expressed circular RNAs in human monocyte derived macrophages response to Mycobacterium tuberculosis infection. Sci Rep 7:13673
Huang Z, Su R, Qing C, Peng Y, Luo Q, Li J (2018a) Plasma circular RNAs hsa_circ_0001953 and hsa_circ_0009024 as diagnostic biomarkers for active tuberculosis. Front Microbiol 9:2010
Huang Z, Su R, Yao F, Peng Y, Luo Q, Li J (2018b) Circulating circular RNAs hsa_circ_0001204 and hsa_circ_0001747 act as diagnostic biomarkers for active tuberculosis detection. Int J Clin Exp Pathol 11:586–594
Huang Z-K, Yao F-Y, Xu J-Q, Deng Z, Su R-G, Peng Y-P, Luo Q, Li J-M (2018c) Microarray expression profile of circular RNAs in peripheral blood mononuclear cells from active tuberculosis patients. Cell Physiol Biochem 45:1230–1240
Huang A, Zheng H, Wu Z, Chen M, Huang Y (2020a) Circular RNA–protein interactions: functions, mechanisms, and identification. Theranostics 10:3503–3517
Huang XY, Huang ZL, Huang J, Xu B, Huang XY, Xu YH, Zhou J, Tang ZY (2020b) Exosomal circRNA-100338 promotes hepatocellular carcinoma metastasis via enhancing invasiveness and angiogenesis. J Exp Clin Cancer Res 39:20
Iparraguirre L, Muñoz-Culla M, Prada-Luengo I, Castillo-Triviño T, Olascoaga J, Otaegui D (2017) Circular RNA profiling reveals that circular RNAs from ANXA2 can be used as new biomarkers for multiple sclerosis. Hum Mol Genet 26:3564–3572
Ivanov A, Memczak S, Wyler E, Torti F, Porath HT, Orejuela MR, Piechotta M, Levanon EY, Landthaler M, Dieterich C et al (2014) Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep 10:170–177
Jeck WR, Sharpless NE (2014) Detecting and characterizing circular RNAs. Nat Biotechnol 32:453–461
Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE (2013) Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 19:141–157
Ji P, Wu W, Chen S, Zheng Y, Zhou L, Zhang J, Cheng H, Yan J, Zhang S, Yang P et al (2019) Expanded expression landscape and prioritization of circular RNAs in mammals. Cell Rep 26:3444-3460.e3445
Jiang M, Lash GE, Zhao X, Long Y, Guo C, Yang H (2018) CircRNA-0004904, circRNA-0001855, and PAPP-A: potential novel biomarkers for the prediction of preeclampsia. Cell Physiol Biochem 46:2576–2586
Joosse SA, Pantel K (2015) Tumor-educated platelets as liquid biopsy in cancer patients. Cancer Cell 28:552–554
Jost I, Shalamova LA, Gerresheim GK, Niepmann M, Bindereif A, Rossbach O (2018) Functional sequestration of microRNA-122 from Hepatitis C Virus by circular RNA sponges. RNA Biol 15(8):1–8
Karczewski KJ, Snyder MP (2018) Integrative omics for health and disease. Nat Rev Genet 19:299–310
Kelly S, Greenman C, Cook PR, Papantonis A (2015) Exon skipping is correlated with exon circularization. J Mol Biol 427:2414–2417
Khan M, Reckman YJ, Aufiero S, van den Hoogenhof M, van der Made I, Beqqali A, Koolbergen DR, Rasmussen TB, van der Velden J, Creemers EE et al (2016) RBM20 regulates circular RNA production from the Titin gene. Circ Res 119:996–1003
Kleaveland B, Shi CY, Stefano J, Bartel DP (2018) A network of noncoding regulatory RNAs acts in the mammalian brain. Cell 174(2):350–362
Kölling M, Haddad G, Wegmann U, Kistler A, Bosakova A, Seeger H, Hübel K, Haller H, Mueller T, Wüthrich RP et al (2019) Circular RNAs in urine of kidney transplant patients with acute T cell-mediated allograft rejection. Clin Chem 65:1287–1294
Kong S, Yang Q, Tang C, Wang T, Shen X, Ju S (2019) Identification of hsa_circ_0001821 as a novel diagnostic biomarker in gastric cancer via comprehensive circular RNA profiling. Front Genet 10:878
Kossenkov AV, Qureshi R, Dawany NB, Wickramasinghe J, Liu Q, Majumdar SR, Chang C, Widura S, Kumar T, Horng W-H et al (2018) A gene expression classifier from whole blood distinguishes benign from malignant lung nodules detected by low-dose CT. Cancer Res 79:263–273
Kramer MC, Liang D, Tatomer DC, Gold B, March ZM, Cherry S, Wilusz JE (2015) Combinatorial control of Drosophila circular RNA expression by intronic repeats, hnRNPs, and SR proteins. Genes Dev 29(20):2168–2182
Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J (2019) The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet 20:1–17
Kumar L, Shamsuzzama HR, Haque R, Baghel T, Nazir A (2017) Circular RNAs: the emerging class of non-coding RNAs and their potential role in human neurodegenerative diseases. Mol Neurobiol 54:7224–7234
Kwapisz D (2017) The first liquid biopsy test approved. Is it a new era of mutation testing for non-small cell lung cancer? Ann Transl Med 5:46
Lam WKJ, Lo YMD (2019) Circular RNAs as urinary biomarkers. Clin Chem 65:1196–1198
Lamb YN, Dhillon S (2017) Epi proColon® 2.0 CE: a blood-based screening test for colorectal cancer. Mol Diagn Ther 21:225–232
Lasda E, Parker R (2016) Circular RNAs co-precipitate with extracellular vesicles: a possible mechanism for circRNA clearance. PLoS ONE 11:e0148407
Legnini I, Timoteo GD, Rossi F, Morlando M, Briganti F, Sthandier O, Fatica A, Santini T, Andronache A, Wade M et al (2017) Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell 66:22–37
Lei B, Zhou J, Xuan X, Tian Z, Zhang M, Gao W, Lin Y, Ni B, Pang H, Fan W (2019) Circular RNA expression profiles of peripheral blood mononuclear cells in hepatocellular carcinoma patients by sequence analysis. Cancer Med 8:1423–1433
Lesko LJ, Atkinson AJ (2001) Use of biomarkers and surrogate endpoints in drug development and regulatory decision making: criteria, validation, strategies. Annu Rev Pharmacol Toxicol 41:347–366
Li P, Chen S, Chen H, Mo X, Li T, Shao Y, Xiao B, Guo J (2015a) Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clin Chim Acta 444:132–136
Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, Chen D, Gu J, He X, Huang S (2015b) Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res 25:981–984
Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Zhong G, Yu B, Hu W, Dai L et al (2015c) Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol 22:256–264
Li M, Xie X, Zhou J, Sheng M, Yin X, Ko E-A, Zhou T, Gu W (2017a) Quantifying circular RNA expression from RNA-seq data using model-based framework. Bioinformatics 33(14):2131–2139
Li S, Li Y, Chen B, Zhao J, Yu S, Tang Y, Zheng Q, Li Y, Wang P, He X et al (2017b) exoRBase: a database of circRNA, lncRNA and mRNA in human blood exosomes. Nucleic Acids Res 46:D106–D112
Li W, Zhong C, Jiao J, Li P, Cui B, Ji C, Ma D (2017c) Characterization of hsa_circ_0004277 as a new biomarker for acute myeloid leukemia via circular RNA profile and bioinformatics analysis. Int J Mol Sci 18(3):597
Li WH, Song YC, Zhang H, Zhou ZJ, Xie X, Zeng QN, Guo K, Wang T, Xia P, Chang DM (2017d) Decreased expression of hsa_circ_00001649 in gastric cancer and its clinical significance. Dis Markers 2017:4587698
Li X, Liu C-X, Xue W, Zhang Y, Jiang S, Yin Q-F, Wei J, Yao R-W, Yang L, Chen L-L (2017e) Coordinated circRNA biogenesis and function with NF90/NF110 in viral infection. Mol Cell 67:214-227.e217
Li J, Li Z, Jiang P, Peng M, Zhang X, Chen K, Liu H, Bi H, Liu X, Li X (2018a) Circular RNA IARS (circ-IARS) secreted by pancreatic cancer cells and located within exosomes regulates endothelial monolayer permeability to promote tumor metastasis. J Exp Clin Cancer Res 37:177
Li L, Li W, Chen N, Zhao H, Xu G, Zhao Y, Pan X, Zhang X, Zhou L, Yu D et al (2018b) FLI1 exonic circular RNAs as a novel oncogenic driver to promote tumor metastasis in small cell lung cancer. Clin Cancer Res 25:1302–1317
Li T, Shao Y, Fu L, Xie Y, Zhu L, Sun W, Yu R, Xiao B, Guo J (2018c) Plasma circular RNA profiling of patients with gastric cancer and their droplet digital RT-PCR detection. J Mol Med (Berl) 96:85–96
Li X, Yang L, Chen L-L (2018d) The biogenesis, functions, and challenges of circular RNAs. Mol Cell 71:428–442
Li Z, Yanfang W, Li J, Jiang P, Peng T, Chen K, Zhao X, Zhang Y, Zhen P, Zhu J et al (2018e) Tumor-released exosomal circular RNA PDE8A promotes invasive growth via the miR-338/MACC1/MET pathway in pancreatic cancer. Cancer Lett 432:237–250
Li Q, Wang Y, Wu S, Zhou Z, Ding X, Shi R, Thorne RF, Zhang XD, Hu W, Wu M (2019a) CircACC1 regulates assembly and activation of AMPK complex under metabolic stress. Cell Metab 30:157–173
Li S, Zhang J, Tan X, Deng J, Li Y, Piao Y, Li C, Yang W, Mo W, Sun J et al (2019b) Microarray expression profile of circular RNAs and mRNAs in children with systemic lupus erythematosus. Clin Rheumatol 38:1339–1350
Li XN, Wang ZJ, Ye CX, Zhao BC, Huang XX, Yang L (2019c) Circular RNA circVAPA is up-regulated and exerts oncogenic properties by sponging miR-101 in colorectal cancer. Biomed Pharmacother 112:108611
Li Y, Zhao J, Yu S, Wang Z, He X, Su Y, Guo T, Sheng H, Chen J, Zheng Q et al (2019d) Extracellular vesicles long RNA sequencing reveals abundant mRNA, circRNA, and lncRNA in human blood as potential biomarkers for cancer diagnosis. Clin Chem 65:798–808
Li Z, Zhou Y, Yang G, He S, Qiu X, Zhang L, Deng Q, Zheng F (2019e) Using circular RNA SMARCA5 as a potential novel biomarker for hepatocellular carcinoma. Clin Chim Acta 492:37–44
Li Z, Zhu X, Huang S (2020) Extracellular vesicle long non-coding RNAs and circular RNAs: biology, functions and applications in cancer. Cancer Lett 489:111–120
Liang D, Wilusz JE (2014) Short intronic repeat sequences facilitate circular RNA production. Genes Dev 28:2233–2247
Liang D, Tatomer DC, Luo Z, Wu H, Yang L, Chen L-L, Cherry S, Wilusz JE (2017) The output of protein-coding genes shifts to circular RNAs when the pre-mRNA processing machinery is limiting. Mol Cell 68:940-954.e943
Liang B, Li M, Deng Q, Wang C, Rong J, He S, Xiang Y, Zheng F (2020) CircRNA ZNF609 in peripheral blood leukocytes acts as a protective factor and a potential biomarker for coronary artery disease. Ann Transl Med 8:741
Lin J, Cai D, Li W, Yu T, Mao H, Jiang S, Xiao B (2019) Plasma circular RNA panel acts as a novel diagnostic biomarker for colorectal cancer. Clin Biochem 74:60–68
Liu C-X, Li X, Nan F, Jiang S, Gao X, Guo S-K, Xue W, Cui Y, Dong K, Ding H et al (2019a) Structure and degradation of circular RNAs regulate PKR activation in innate immunity. Cell 177(4):865–880
Liu XX, Yang YE, Liu X, Zhang MY, Li R, Yin YH, Qu YQ (2019b) A two-circular RNA signature as a noninvasive diagnostic biomarker for lung adenocarcinoma. J Transl Med 17:50
Liu YT, Han XH, Xing PY, Hu XS, Hao XZ, Wang Y, Li JL, Zhang ZS, Yang ZH, Shi YK (2019c) Circular RNA profiling identified as a biomarker for predicting the efficacy of Gefitinib therapy for non-small cell lung cancer. J Thorac Dis 11:1779–1787
Liu H, Lu G, Wang W, Jiang X, Gu S, Wang J, Yan X, He F, Wang J (2020) A panel of circRNAs in the serum serves as biomarkers for Mycobacterium tuberculosis Infection. Front Microbiol 11:1215
Loon EV, Gazut S, Yazdani S, Lerut E, de Loor H, Coemans M, Noël L-H, Thorrez L, Lommel LV, Schuit F et al (2019) Development and validation of a peripheral blood mRNA assay for the assessment of antibody-mediated kidney allograft rejection: a multicentre, prospective study. EBioMedicine 46:463–472
Lu J, Zhang PY, Xie JW, Wang JB, Lin JX, Chen QY, Cao LL, Huang CM, Li P, Zheng CH (2019a) hsa_circ_0000467 promotes cancer progression and serves as a diagnostic and prognostic biomarker for gastric cancer. J Clin Lab Anal 33:e22726
Lu J, Zhang PY, Xie JW, Wang JB, Lin JX, Chen QY, Cao LL, Li P, Zheng CH, Huang CM (2019b) Circular RNA hsa_circ_0006848 related to ribosomal protein L6 acts as a novel biomarker for early gastric cancer. Dis Markers 2019:3863458
Lu X, Song M, Wang F (2019c) Circulating circular RNAs as biomarkers of cancer. Non-coding RNA Investig 3:8
Lukiw WJ (2013) Circular RNA (circRNA) in Alzheimer’s disease (AD). Front Genet 4:307
Luo Q, Zhang L, Li X, Fu B, Deng Z, Qing C, Su R, Xu J, Guo Y, Huang Z et al (2018) Identification of circular RNAs hsa_circ_0044235 in peripheral blood as novel biomarkers for rheumatoid arthritis. Clin Exp Immunol 194:118–124
Luo Q, Zhang L, Li X, Fu B, Guo Y, Huang Z, Li J (2019) Identification of circular RNAs hsa_circ_0044235 and hsa_circ_0068367 as novel biomarkers for systemic lupus erythematosus. Int J Mol Med 44:1462–1472
Luo H, Zhao Q, Wei W, Zheng L, Yi S, Li G, Wang W, Sheng H, Pu H, Mo H et al (2020a) Circulating tumor DNA methylation profiles enable early diagnosis, prognosis prediction, and screening for colorectal cancer. Sci Transl Med 12:eaax7533
Luo HL, Peng Y, Luo H, Zhang JA, Liu GB, Xu H, Huang GX, Sun YF, Huang J, Zheng BY et al (2020b) Circular RNA hsa_circ_0001380 in peripheral blood as a potential diagnostic biomarker for active pulmonary tuberculosis. Mol Med Rep 21:1890–1896
Luo Y-H, Yang Y-P, Chien C-S, Yarmishyn AA, Ishola AA, Chien Y, Chen Y-M, Huang T-W, Lee K-Y, Huang W-C et al (2020c) Plasma level of circular RNA hsa_circ_0000190 correlates with tumor progression and poor treatment response in advanced lung cancers. Cancers 12:1740
Ma J, Qi G, Li L (2020) A novel serum exosomes-based biomarker hsa_circ_0002130 facilitates osimertinib-resistance in non-small cell lung cancer by sponging miR-498. Onco Targets Ther 13:5293–5307
Maas SLN, Breakefield XO, Weaver AM (2017) Extracellular vesicles: unique intercellular delivery vehicles. Trends Cell Biol 27:172–188
Maass PG, Glažar P, Memczak S, Dittmar G, Hollfinger I, Schreyer L, Sauer AV, Toka O, Aiuti A, Luft FC et al (2017) A map of human circular RNAs in clinically relevant tissues. J Mol Med (Berl) 95:1179–1189
MacLean E, Broger T, Yerlikaya S, Fernandez-Carballo BL, Pai M, Denkinger CM (2019) A systematic review of biomarkers to detect active tuberculosis. Nat Microbiol 4:748–758
Marshall K, Mohr S, Khettabi F, Nossova N, Chao S, Bao W, Ma J, Li XJ, Liew CC (2010) A blood-based biomarker panel for stratifying current risk for colorectal cancer. Int J Cancer 126:1177–1186
Mattox AK, Bettegowda C, Zhou S, Papadopoulos N, Kinzler KW, Vogelstein B (2019) Applications of liquid biopsies for cancer. Sci Transl Med 11:eaay1984
Max KEA, Bertram K, Akat K, Bogardus KA, Li J, Morozov P, Ben-Dov IZ, Li X, Weiss ZR, Azizian A et al (2018) Human plasma and serum extracellular small RNA reference profiles and their clinical utility. Proc Natl Acad Sci USA 115:201714397
Mayhew MB, Buturovic L, Luethy R, Midic U, Moore AR, Roque JA, Shaller BD, Asuni T, Rawling D, Remmel M et al (2020) A generalizable 29-mRNA neural-network classifier for acute bacterial and viral infections. Nat Commun 11:1177
Mei M, Wang Y, Wang Q, Liu Y, Song W, Zhang M (2019) CircCDYL serves as a new biomarker in Mantle cell lymphoma and promotes cell proliferation. Cancer Manag Res 11:10215–10221
Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M et al (2013) Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495:333–338
Memczak S, Papavasileiou P, Peters O, Rajewsky N (2015) Identification and characterization of circular RNAs as a new class of putative biomarkers in human blood. PLoS ONE 10:e0141214
Meng S, Zhou H, Feng Z, Xu Z, Tang Y, Li P, Wu M (2017) CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer 16:94
Miao Q, Zhong Z, Jiang Z, Lin Y, Ni B, Yang W, Tang J (2019) RNA-seq of circular RNAs identified circPTPN22 as a potential new activity indicator in systemic lupus erythematosus. Lupus 28:520–528
Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O’Briant KC, Allen A et al (2008) Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci USA 105:10513–10518
Ng WL, Marinov GK, Liau ES, Lam YL, Lim Y-Y, Ea C-K (2016) Inducible RasGEF1B circular RNA is a positive regulator of ICAM-1 in the TLR4/LPS pathway. RNA Biol 13:861–871
Nicolet BP, Engels S, Aglialoro F, van den Akker E, von Lindern M, Wolkers MC (2018) Circular RNA expression in human hematopoietic cells is widespread and cell-type specific. Nucleic Acids Res 46:8168–8180
Ouyang Q, Huang Q, Jiang Z, Zhao J, Shi GP, Yang M (2018) Using plasma circRNA_002453 as a novel biomarker in the diagnosis of lupus nephritis. Mol Immunol 101:531–538
Ouyang Q, Wu J, Jiang Z, Zhao J, Wang R, Lou A, Zhu D, Shi GP, Yang M (2017) Microarray expression profile of circular RNAs in peripheral blood mononuclear cells from rheumatoid arthritis patients. Cell Physiol Biochem 42:651–659
Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L, Hanan M, Wyler E, Perez-Hernandez D, Ramberger E et al (2017) Translation of circRNAs. Mol Cell 66:9–21
Pan B, Qin J, Liu X, He B, Wang X, Pan Y, Sun H, Xu T, Xu M, Chen X et al (2019) Identification of serum exosomal hsa-circ-0004771 as a novel diagnostic biomarker of colorectal cancer. Front Genet 10:1096
Pardini B, Sabo A, Birolo G, Calin G (2019) Noncoding RNAs in extracellular fluids as cancer biomarkers: the new frontier of liquid biopsies. Cancers 11:1170
Park O, Ha H, Lee Y, Boo S, Kwon D, Song H, Kim Y (2019) Endoribonucleolytic cleavage of m6A-containing RNAs by RNase P/MRP complex. Mol Cell 74(3):494–507
Peng YK, Pu K, Su HX, Zhang J, Zheng Y, Ji R, Guo QH, Wang YP, Guan QL, Zhou YN (2020) Circular RNA hsa_circ_0010882 promotes the progression of gastric cancer via regulation of the PI3K/Akt/mTOR signaling pathway. Eur Rev Med Pharmacol Sci 24:1142–1151
Ping L, Jian-Jun C, Chu-Shu L, Guang-Hua L, Ming Z (2019) High circ_100053 predicts a poor outcome for chronic myeloid leukemia and is involved in imatinib resistance. Oncol Res. https://doi.org/10.3727/096504018x15412701483326
Piwecka M, Glažar P, Hernandez-Miranda LR, Memczak S, Wolf SA, Rybak-Wolf A, Filipchyk A, Klironomos F, Cerda Jara CA, Fenske P et al (2017) Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science 357:eaam8526
Powers KA, Ghani AC, Miller WC, Hoffman IF, Pettifor AE, Kamanga G, Martinson FE, Cohen MS (2011) The role of acute and early HIV infection in the spread of HIV and implications for transmission prevention strategies in Lilongwe, Malawi: a modelling study. Lancet 378:256–268
Preußer C, Hung L-H, Schneider T, Schreiner S, Hardt M, Moebus A, Santoso S, Bindereif A (2018) Selective release of circRNAs in platelet-derived extracellular vesicles. J Extracell Vesicles 7:1424473
Qian Z, Liu H, Li M, Shi J, Li N, Zhang Y, Zhang X, Lv J, Xie X, Bai Y et al (2018) Potential diagnostic power of blood circular RNA expression in active pulmonary tuberculosis. EBioMedicine 27:18–26
Qian Z, Lv J, Kelly GT, Wang H, Zhang X, Gu W, Yin X, Wang T, Zhou T (2016) Expression of nuclear factor, erythroid 2-like 2-mediated genes differentiates tuberculosis. Tuberculosis 99:56–62
Qiao GL, Chen L, Jiang WH, Yang C, Yang CM, Song LN, Chen Y, Yan HL, Ma LJ (2019) Hsa_circ_0003998 may be used as a new biomarker for the diagnosis and prognosis of hepatocellular carcinoma. Onco Targets Ther 12:5849–5860
Ramilo O, Mejias A (2017) Host transcriptomics for diagnosis of infectious diseases: one step closer to clinical application. Eur Respir J 49:1700993
Ravnik-Glavač M, Glavač D (2020) Circulating RNAs as potential biomarkers in amyotrophic lateral sclerosis. Int J Mol Sci 21:1714
Revenfeld ALS, Bæk R, Nielsen MH, Stensballe A, Varming K, Jørgensen M (2014) Diagnostic and prognostic potential of extracellular vesicles in peripheral blood. Clin Ther 36:830–846
Richey LE, Halperin J (2013) Acute human immunodeficiency virus infection. Am J Med Sci 345:136–142
Rong D, Lu C, Zhang B, Fu K, Zhao S, Tang W, Cao H (2019) CircPSMC3 suppresses the proliferation and metastasis of gastric cancer by acting as a competitive endogenous RNA through sponging miR-296-5p. Mol Cancer 18:25
Rose S, Contrepois K, Moneghetti KJ, Zhou W, Mishra T, Mataraso S, Dagan-Rosenfeld O, Ganz AB, Dunn J, Hornburg D et al (2019) A longitudinal big data approach for precision health. Nat Med 25:792–804
Ross RW, Galsky MD, Scher HI, Magidson J, Wassmann K, Lee G-S, Katz L, Subudhi SK, Anand A, Fleisher M et al (2012) A whole-blood RNA transcript-based prognostic model in men with castration-resistant prostate cancer: a prospective study. Lancet Oncol 13:1105–1113
Rubis G, Krishnan S, Bebawy M (2019) Liquid biopsies in cancer diagnosis, monitoring, and prognosis. Trends Pharmacol Sci 40:172–186
Rybak-Wolf A, Stottmeister C, Glažar P, Jens M, Pino N, Giusti S, Hanan M, Behm M, Bartok O, Ashwal-Fluss R et al (2015) Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell 58:870–885
Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO (2013) Cell-type specific features of circular RNA expression. PLoS Genet 9(9):e1003777
Salzman J, Gawad C, Wang P, Lacayo N, Brown PO (2012) Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE. https://doi.org/10.1371/journal.pone.0030733
Sambarey A, Devaprasad A, Mohan A, Ahmed A, Nayak S, Swaminathan S, D’Souza G, Jesuraj A, Dhar C, Babu S et al (2017) Unbiased identification of blood-based biomarkers for pulmonary tuberculosis by modeling and mining molecular interaction networks. EBioMedicine 15:112–126
Sanger HL, Klotz G, Riesner D, Gross HJ, Kleinschmidt AK (1976) Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc Natl Acad Sci USA 73:3852–3856
Schnell A, Schmidl C, Herr W, Siska PJ (2018) The peripheral and intratumoral immune cell landscape in cancer patients: a proxy for tumor biology and a tool for outcome prediction. Biomedicines 6:25
Shaked Y (2019) The pro-tumorigenic host response to cancer therapies. Nat Rev Cancer 19:667–685
Shang A, Gu C, Wang W, Wang X, Sun J, Zeng B, Chen C, Chang W, Ping Y, Ji P et al (2020) Exosomal circPACRGL promotes progression of colorectal cancer via the miR-142-3p/miR-506-3p-TGF-β1 axis. Mol Cancer 19:117
Shang Q, Yang Z, Jia R, Ge S (2019) The novel roles of circRNAs in human cancer. Mol Cancer 18:6
Shao Y, Tao X, Lu R, Zhang H, Ge J, Xiao B, Ye G, Guo J (2020) Hsa_circ_0065149 is an indicator for early gastric cancer screening and prognosis prediction. Pathol Oncol Res 26:1475–1482
Sheng R, Li X, Wang Z, Wang X (2020) Circular RNAs and their emerging roles as diagnostic and prognostic biomarkers in ovarian cancer. Cancer Lett 473:139–147
Shi L, Tao C, Tang Y, Xia Y, Li X, Wang X (2020) Hypoxia-induced hsa_circ_0000826 is linked to liver metastasis of colorectal cancer. J Clin Lab Anal 34(9):e23405
Showe MK, Vachani A, Kossenkov AV, Yousef M, Nichols C, Nikonova EV, Chang C, Kucharczuk J, Tran B, Wakeam E et al (2009) Gene expression profiles in peripheral blood mononuclear cells can distinguish patients with non-small cell lung cancer from patients with nonmalignant lung disease. Cancer Res 69:9202–9210
Shuai M, Hong J, Huang D, Zhang X, Tian Y (2018) Upregulation of circRNA_0000285 serves as a prognostic biomarker for nasopharyngeal carcinoma and is involved in radiosensitivity. Oncol Lett 16:6495–6501
Siravegna G, Marsoni S, Siena S, Bardelli A (2017) Integrating liquid biopsies into the management of cancer. Nat Rev Clin Oncol 14:531–548
Smid M, Wilting SM, Uhr K, Rodríguez-González GF, de Weerd V, van der Smissen WJC, van der Vlugt-Daane M, van Galen A, Nik-Zainal S, Butler A et al (2019) The circular RNome of primary breast cancer. Genome Res 29:356–366
Sole C, Arnaiz E, Manterola L, Otaegui D, Lawrie CH (2019) The circulating transcriptome as a source of cancer liquid biopsy biomarkers. Semin Cancer Biol 58:100–108
Song R, Bai Y, Li X, Zhu J, Zhang H, Shi Y, Li K, Wang B, Zhang H, Yang Y et al (2020) Plasma circular RNA DYM related to major depressive disorder and rapid antidepressant effect treated by visual cortical repetitive transcranial magnetic stimulation. J Affect Disord 274:486–493
Sonnenschein K, Wilczek A, de Gonzalo-Calvo D, Pfanne A, Derda A, Zwadlo C, Bavendiek U, Bauersachs J, Fiedler J, Thum T (2019) Serum circular RNAs act as blood-based biomarkers for hypertrophic obstructive cardiomyopathy. Sci Rep 9:20350
Stark R, Grzelak M, Hadfield J (2019) RNA sequencing: the teenage years. Nat Rev Genet 20:1–26
Starke S, Jost I, Rossbach O, Schneider T, Schreiner S, Hung L-H, Bindereif A (2014) Exon circularization requires canonical splice signals. Cell Rep 10:103–111
Su M, Xiao Y, Ma J, Tang Y, Tian B, Zhang Y, Li X, Wu Z, Yang D, Zhou Y et al (2019) Circular RNAs in cancer: emerging functions in hallmarks, stemness, resistance and roles as potential biomarkers. Mol Cancer 18:90
Sun J-Y, Shi Y, Cai X-Y, Liu J (2020) Potential diagnostic and therapeutic value of circular RNAs in cardiovascular diseases. Cell Signal 71:109604
Sun X-H, Wang Y-T, Li G-F, Zhang N, Fan L (2020) Serum-derived three-circRNA signature as a diagnostic biomarker for hepatocellular carcinoma. Cancer Cell Int 20:226
Sun Y, Jiang X, Lv Y, Liang X, Zhao B, Bian W, Zhang D, Jiang J, Zhang C (2020) Circular RNA expression profiles in plasma from patients with heart failure related to platelet activity. Biomolecules 10(2):187
Surinova S, Radová L, Choi M, Srovnal J, Brenner H, Vitek O, Hajdúch M, Aebersold R (2015) Non-invasive prognostic protein biomarker signatures associated with colorectal cancer. EMBO Mol Med 7:1153–1165
Sweeney TE, Perumal TM, Henao R, Nichols M, Howrylak JA, Choi AM, Bermejo-Martin JFF, Almansa R, Tamayo E, Davenport EE et al (2018) A community approach to mortality prediction in sepsis via gene expression analysis. Nat Commun 9:694
Szabo L, Morey R, Palpant NJ, Wang PL, Afari N, Jiang C, Parast MM, Murry CE, Laurent LC, Salzman J (2015) Statistically based splicing detection reveals neural enrichment and tissue-specific induction of circular RNA during human fetal development. Genome Biol 16:126
Szabo L, Salzman J (2016) Detecting circular RNAs: bioinformatic and experimental challenges. Nat Rev Genet 17:679–692
Tan S, Gou Q, Pu W, Guo C, Yang Y, Wu K, Liu Y, Liu L, Wei Y-Q, Peng Y (2018) Circular RNA F-circEA produced from EML4-ALK fusion gene as a novel liquid biopsy biomarker for non-small cell lung cancer. Cell Res 28:693–695
Tang W, Fu K, Sun H, Rong D, Wang H, Cao H (2018) CircRNA microarray profiling identifies a novel circulating biomarker for detection of gastric cancer. Mol Cancer 17:137
Tang X, Liu S, Ding Y, Guo C, Guo J, Hua K, Qiu J (2020) Serum circular FoxO3a serves as a novel prognostic biomarker in squamous cervical cancer. Cancer Manag Res 12:2531–2540
Tao X, Shao Y, Lu R, Ye Q, Xiao B, Ye G, Guo J (2020) Clinical significance of hsa_circ_0000419 in gastric cancer screening and prognosis estimation. Pathol Res Pract 216:152763
Teng L, Chen Y, Chen H, He X, Wang J, Peng Y, Duan H, Li H, Lin D, Shao B (2017) Circular RNA hsa_circ_0021001 in peripheral blood: a potential novel biomarker in the screening of intracranial aneurysm. Oncotarget 8:107125–107133
Torrano V, Royo F, Peinado H, Loizaga-Iriarte A, Unda M, Falcón-Perez JM, Carracedo A (2016) Vesicle-MaNiA: extracellular vesicles in liquid biopsy and cancer. Curr Opin Pharmacol 29:47–53
Tsalik EL, Henao R, Nichols M, Burke T, Ko ER, McClain MT, Hudson LL, Mazur A, Freeman DH, Veldman T et al (2016) Host gene expression classifiers diagnose acute respiratory illness etiology. Sci Transl Med 8:322ra311
Turchinovich A, Weiz L, Langheinz A, Burwinkel B (2011) Characterization of extracellular circulating microRNA. Nucleic Acids Res 39:7223–7233
Valk PJM, Verhaak RGW, Beijen MA, Erpelinck CAJ, van Doorn-Khosrovani SBvW, Boer JM, Beverloo HB, Moorhouse MJ, van der Spek PJ, Löwenberg B et al (2004) Prognostically useful gene-expression profiles in acute myeloid leukemia. N Engl J Med 350:1617–1628
van Heesch S, Witte F, Schneider-Lunitz V, Schulz JF, Adami E, Faber AB, Kirchner M, Maatz H, Blachut S, Sandmann C-L et al (2019) The translational landscape of the human heart. Cell 178(1):242–260
Vargas AJ, Harris CC (2016) Biomarker development in the precision medicine era: lung cancer as a case study. Nat Rev Cancer 16:525–537
Vargas J, Lima JA, Kraus WE, Douglas PS, Rosenberg S (2013) Use of the Corus® CAD gene expression test for assessment of obstructive coronary artery disease likelihood in symptomatic non-diabetic patients. PLoS Curr. https://doi.org/10.1371/currents.eogt.0f04f6081905998fa92b99593478aeab
Vausort M, Salgado-Somoza A, Zhang L, Leszek P, Scholz M, Teren A, Burkhardt R, Thiery J, Wagner DR, Devaux Y (2016) Myocardial infarction-associated circular RNA predicting left ventricular dysfunction. J Am Coll Cardiol 68:1247–1248
Venø MT, Hansen TB, Venø ST, Clausen BH, Grebing M, Finsen B, Holm IE, Kjems J (2015) Spatio-temporal regulation of circular RNA expression during porcine embryonic brain development. Genome Biol 16:245
Vicens Q, Westhof E (2014) Biogenesis of circular RNAs. Cell 159:13–14
Vo JN, Cieslik M, Zhang Y, Shukla S, Xiao L, Zhang Y, Wu Y-M, Dhanasekaran SM, Engelke CG, Cao X et al (2019) The landscape of circular RNA in cancer. Cell 176:869-881.e813
Wan JCM, Massie C, Garcia-Corbacho J, Mouliere F, Brenton JD, Caldas C, Pacey S, Baird R, Rosenfeld N (2017) Liquid biopsies come of age: towards implementation of circulating tumour DNA. Nat Rev Cancer 17:223–238
Wang Y, Wang Z (2015) Efficient backsplicing produces translatable circular mRNAs. RNA 21:172–179
Wang PL, Bao Y, Yee M-C, Barrett SP, Hogan GJ, Olsen MN, Dinneny JR, Brown PO, Salzman J (2014) Circular RNA is expressed across the eukaryotic tree of life. PLoS ONE 9(6):e90859
Wang YM, Huang LM, Li DR, Shao JH, Xiong SL, Wang CM, Lu SM (2017a) Hsa_circ_0101996 combined with hsa_circ_0101119 in peripheral whole blood can serve as the potential biomarkers for human cervical squamous cell carcinoma. Int J Clin Exp Pathol 10:11924–11931
Wang Z-Y, Guo Z-D, Li J-M, Zhao Z-Z, Fu Y-Y, Zhang C-M, Zhang Y, Liu L-N, Qian J, Liu L-N (2017b) Genome-wide search for competing endogenous RNAs responsible for the effects induced by Ebola virus replication and transcription using a trVLP system. Front Cell Infect Microbiol 7:479
Wang L, Shen C, Wang Y, Zou T, Zhu H, Lu X, Li L, Yang B, Chen J, Chen S et al (2019a) Identification of circular RNA hsa_circ_0001879 and hsa_circ_0004104 as novel biomarkers for coronary artery disease. Atherosclerosis 286:88–96
Wang Q, Zhang Q, Sun H, Tang W, Yang L, Xu Z, Liu Z, Jin H, Cao X (2019b) Circ-TTC17 promotes proliferation and migration of esophageal squamous cell carcinoma. Dig Dis Sci 64:751–758
Wang W, Dong R, Guo Y, He J, Shao C, Yi P, Yu F, Gu D, Zheng J (2019c) CircMTO1 inhibits liver fibrosis via regulation of miR-17-5p and Smad7. J Cell Mol Med 23:5486–5496
Wang W, Wang J, Zhang X, Liu G (2019d) Serum circSETDB1 is a promising biomarker for predicting response to platinum-taxane-combined chemotherapy and relapse in high-grade serous ovarian cancer. Onco Targets Ther 12:7451–7457
Wang Y, Liu J, Ma J, Sun T, Zhou Q, Wang W, Wang G, Wu P, Wang H, Jiang L et al (2019e) Exosomal circRNAs: biogenesis, effect and application in human diseases. Mol Cancer 18:116
Wang Y, Wu C, Zhang F, Zhang Y, Ren Z, Lammi MJ, Guo X (2019f) Screening for differentially expressed circular RNAs in the cartilage of osteoarthritis patients for their diagnostic value. Genet Test Mol Biomark 23:706–716
Wang J, Kong J, Nie Z, Chen D, Qiang J, Gao W, Chen X (2020a) Circular RNA hsa_circ_0066755 as an oncogene via sponging miR-651 and as a promising diagnostic biomarker for nasopharyngeal carcinoma. Int J Med Sci 17:1499–1507
Wang J, Zhao X, Wang Y, Ren F, Sun D, Yan Y, Kong X, Bu J, Liu M, Xu S (2020b) CircRNA-002178 act as a ceRNA to promote PDL1/PD1 expression in lung adenocarcinoma. Cell Death Dis 11:32
Wang Q, Liu H, Liu Z, Yang L, Zhou J, Cao X, Sun H (2020c) Circ-SLC7A5, a potential prognostic circulating biomarker for detection of ESCC. Cancer Genet 240:33–39
Wang W, Li Y, Li X, Liu B, Han S, Li X, Zhang B, Li J, Sun S (2020d) Circular RNA circ-FOXP1 induced by SOX9 promotes hepatocellular carcinoma progression via sponging miR-875-3p and miR-421. Biomed Pharmacother 121:109517
Wang Y, Wu C, Zhang Y, Yang Y, Ren Z, Lammi MJ, Guo X (2020e) Screening for differentially expressed circRNA between Kashin-Beck disease and osteoarthritis patients based on circRNA chips. Clin Chim Acta 501:92–101
Warsinske H, Vashisht R, Khatri P (2019) Host-response-based gene signatures for tuberculosis diagnosis: a systematic comparison of 16 signatures. PLoS Med 16:e1002786
Weng W, Wei Q, Toden S, Yoshida K, Nagasaka T, Fujiwara T, Cai S, Qin H, Ma Y, Goel A (2017) Circular RNA ciRS-7—a promising prognostic biomarker and a potential therapeutic target in colorectal cancer. Clin Cancer Res 23:3918–3928
Westholm JO, Miura P, Olson S, Shenker S, Joseph B, Sanfilippo P, Celniker SE, Graveley BR, Lai EC (2014) Genome-wide analysis of Drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep 9:1966–1980
Wilusz JE (2015) Repetitive elements regulate circular RNA biogenesis. Mob Genet Elem 5:39–45
Wu H, Yang L, Chen L-L (2017a) The diversity of long noncoding RNAs and their generation. Trends Genet 33:540–552
Wu N, Jin L, Cai J (2017b) Profiling and bioinformatics analyses reveal differential circular RNA expression in hypertensive patients. Clin Exp Hypertens 39:454–459
Wu J, Li J, Liu H, Yin J, Zhang M, Yu Z, Miao H (2019a) Circulating plasma circular RNAs as novel diagnostic biomarkers for congenital heart disease in children. J Clin Lab Anal 33:e22998
Wu J, Zhou Q, Niu Y, Chen J, Zhu Y, Ye S, Xi Y, Wang F, Qiu H, Bu S (2019b) Aberrant expression of serum circANRIL and hsa_circ_0123996 in children with Kawasaki disease. J Clin Lab Anal 33:e22874
Wu W, Ji P, Zhao F (2020a) CircAtlas: an integrated resource of one million highly accurate circular RNAs from 1070 vertebrate transcriptomes. Genome Biol 21:101
Wu WP, Pan YH, Cai MY, Cen JM, Chen C, Zheng L, Liu X, Xiong XD (2020b) Plasma-derived exosomal circular RNA hsa_circ_0005540 as a novel diagnostic biomarker for coronary artery disease. Dis Markers 2020:3178642
Wu Z, Sun H, Liu W, Zhu H, Fu J, Yang C, Fan L, Wang L, Liu Y, Xu W et al (2020c) Circ-RPL15: a plasma circular RNA as novel oncogenic driver to promote progression of chronic lymphocytic leukemia. Leukemia 34:919–923
Xia X, Li X, Li F, Wu X, Zhang M, Zhou H, Huang N, Yang X, Xiao F, Liu D et al (2019) A novel tumor suppressor protein encoded by circular AKT3 RNA inhibits glioblastoma tumorigenicity by competing with active phosphoinositide-dependent Kinase-1. Mol Cancer 18:131
Xiao M-S, Ai Y, Wilusz JE (2020) Biogenesis and functions of circular RNAs come into focus. Trends Cell Biol 30(3):226–240
Xie M, Yu T, Jing X, Ma L, Fan Y, Yang F, Ma P, Jiang H, Wu X, Shu Y et al (2020a) Exosomal circSHKBP1 promotes gastric cancer progression via regulating the miR-582-3p/HUR/VEGF axis and suppressing HSP90 degradation. Mol Cancer 19:112
Xie R, Zhang Y, Zhang J, Li J, Zhou X (2020b) The role of circular RNAs in immune-related diseases. Front Immunol 11:545
Xie Y, Li J, Li P, Li N, Zhang Y, Binang H, Zhao Y, Duan W, Chen Y, Wang Y et al (2020c) RNA-Seq profiling of serum exosomal circular RNAs reveals circ-PNN as a potential biomarker for human colorectal cancer. Front Oncol 10:982
Xiong S, Peng H, Ding X, Wang X, Wang L, Wu C, Wang S, Xu H, Liu Y (2019) Circular RNA expression profiling and the potential role of hsa_circ_0089172 in Hashimoto’s thyroiditis via sponging miR125a-3p. Mol Ther Nucleic Acids 17:38–48
Xu H, Guo S, Li W, Yu P (2015) The circular RNA Cdr1as, via miR-7 and its targets, regulates insulin transcription and secretion in islet cells. Sci Rep 5:12453
Xu H, Gong Z, Shen Y, Fang Y, Zhong S (2018a) Circular RNA expression in extracellular vesicles isolated from serum of patients with endometrial cancer. Epigenomics 10:187–197
Xu Z, Li P, Fan L, Wu M (2018b) The potential role of circRNA in tumor immunity regulation and immunotherapy. Front Immunol 9:9
Xu M, Xie F, Tang X, Wang T, Wang S (2020a) Insights into the role of circular RNA in macrophage activation and fibrosis disease. Pharmacol Res 156:104777
Xu Y, Leng K, Yao Y, Kang P, Liao G, Han Y, Shi G, Ji D, Huang P, Zheng W et al (2020b) A novel circular RNA, circ-CCAC1, contributes to CCA progression, induces angiogenesis, and disrupts vascular endothelial barriers. Hepatology. https://doi.org/10.1002/hep.31493
Yan B, Zhang Y, Liang C, Liu B, Ding F, Wang Y, Zhu B, Zhao R, Yu X-Y, Li Y (2020) Stem cell-derived exosomes prevent pyroptosis and repair ischemic muscle injury through a novel exosome/circHIPK3/FOXO3a pathway. Theranostics 10:6728–6742
Yang F, Liu DY, Guo JT, Ge N, Zhu P, Liu X, Wang S, Wang GX, Sun SY (2017a) Circular RNA circ-LDLRAD3 as a biomarker in diagnosis of pancreatic cancer. World J Gastroenterol 23:8345–8354
Yang Q, Du WW, Wu N, Yang W, Awan F, Fang L, Ma J, Li X, Zeng Y, Yang Z et al (2017b) A circular RNA promotes tumorigenesis by inducing c-myc nuclear translocation. Cell Death Differ 24(9):1609–1620
Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, Jin Y, Yang Y, Chen L, Wang Y et al (2017c) Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res 27:626–641
Yang L, Fu J, Zhou Y (2018) Circular RNAs and their emerging roles in immune regulation. Front Immunol 9:2977
Yang X, Li J, Wu Y, Ni B, Zhang B (2019) Aberrant dysregulated circular RNAs in the peripheral blood mononuclear cells of patients with rheumatoid arthritis revealed by RNA sequencing: novel diagnostic markers for RA. Scand J Clin Lab Investig 79:551–559
Yao G, Niu W, Zhu X, He M, Kong L, Chen S, Zhang L, Cheng Z (2019) hsa_circRNA_104597: a novel potential diagnostic and therapeutic biomarker for schizophrenia. Biomark Med 13:331–340
Ye DX, Wang SS, Huang Y, Chi P (2019) A 3-circular RNA signature as a noninvasive biomarker for diagnosis of colorectal cancer. Cancer Cell Int 19:276
Yi Z, Gao K, Li R, Fu Y (2018) Dysregulated circRNAs in plasma from active tuberculosis patients. J Cell Mol Med 22:4076–4084
Yin WB, Yan MG, Fang X, Guo JJ, Xiong W, Zhang RP (2018) Circulating circular RNA hsa_circ_0001785 acts as a diagnostic biomarker for breast cancer detection. Clin Chim Acta 487:363–368
Yoshimoto R, Rahimi K, Hansen T, Kjems J, Mayeda A (2019) Biosynthesis of circular RNA ciRS-7/CDR1as is mediated by mammalian-wide interspersed repeats (MIRs). Biorxiv 411231
You X, Vlatkovic I, Babic A, Will T, Epstein I, Tushev G, Akbalik G, Wang M, Glock C, Quedenau C (2015) Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat Neurosci 18:603–610
Yu C-Y, Li T-C, Wu Y-Y, Yeh C-H, Chiang W, Chuang C-Y, Kuo H-C (2017) The circular RNA circBIRC6 participates in the molecular circuitry controlling human pluripotency. Nat Commun 8:1149
Yu M, Bardia A, Wittner BS, Stott SL, Smas ME, Ting DT, Isakoff SJ, Ciciliano JC, Wells MN, Shah AM et al (2013) Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science 339:580–584
Zaas AK, Chen M, Varkey J, Veldman T, Hero AO, Lucas J, Huang Y, Turner R, Gilbert A, Lambkin-Williams R et al (2009) Gene expression signatures diagnose Influenza and other symptomatic respiratory viral infections in humans. Cell Host Microbe 6:207–217
Zak DE, Penn-Nicholson A, Scriba TJ, Thompson E, Suliman S, Amon LM, Mahomed H, Erasmus M, Whatney W, Hussey GD et al (2016) A blood RNA signature for tuberculosis disease risk: a prospective cohort study. Lancet 387:2312–2322
Zaporozhchenko IA, Ponomaryova AA, Rykova EY, Laktionov PP (2018) The potential of circulating cell-free RNA as a cancer biomarker: challenges and opportunities. Expert Rev Mol Diagn 18:133–145
Zemmour H, Planer D, Magenheim J, Moss J, Neiman D, Gilon D, Korach A, Glaser B, Shemer R, Landesberg G et al (2018) Non-invasive detection of human cardiomyocyte death using methylation patterns of circulating DNA. Nat Commun 9:1443
Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, Zhu S, Yang L, Chen LL (2013) Circular intronic long noncoding RNAs. Mol Cell 51:792–806
Zhang X-O, Wang H-B, Zhang Y, Lu X, Chen L-L, Yang L (2014) Complementary sequence-mediated exon circularization. Cell 159:134–147
Zhang YG, Yang HL, Long Y, Li WL (2016) Circular RNA in blood corpuscles combined with plasma protein factor for early prediction of pre-eclampsia. BJOG 123:2113–2118
Zhang Y, Li J, Yu J, Liu H, Shen Z, Ye G, Mou T, Qi X, Li G (2017) Circular RNAs signature predicts the early recurrence of stage III gastric cancer after radical surgery. Oncotarget 8:22936–22943
Zhang C, Wang X, Chen Y, Wu Z, Zhang C, Shi W (2018a) The down-regulation of hsa_circ_0012919, the sponge for miR-125a-3p, contributes to DNA methylation of CD11a and CD70 in CD4(+) T cells of systemic lupus erythematous. Clin Sci (Lond) 132:2285–2298
Zhang H, Deng T, Ge S, Liu Y, Bai M, Zhu K, Fan Q, Li J, Ning T, Tian F et al (2018b) Exosome circRNA secreted from adipocytes promotes the growth of hepatocellular carcinoma by targeting deubiquitination-related USP7. Oncogene 38:2844–2859
Zhang J, Xu Y, Xu S, Liu Y, Yu L, Li Z, Xue X, Wang H (2018c) Plasma circular RNAs, hsa_circRNA_025016, predict postoperative atrial fibrillation after isolated off-pump coronary artery bypass grafting. J Am Heart Assoc 7:e006642
Zhang M, Zhao K, Xu X, Yang Y, Yan S, Wei P, Liu H, Xu J, Xiao F, Zhou H et al (2018d) A peptide encoded by circular form of LINC-PINT suppresses oncogenic transcriptional elongation in glioblastoma. Nat Commun 9:4475
Zhang MY, Wang JB, Zhu ZW, Li LJ, Liu RS, Yang XK, Leng RX, Li XM, Pan HF, Ye DQ (2018e) Differentially expressed circular RNAs in systemic lupus erythematosus and their clinical significance. Biomed Pharmacother 107:1720–1727
Zhang W, Yang S, Liu Y, Wang Y, Lin T, Li Y, Zhang R (2018f) Hsa_circ_0007534 as a blood-based marker for the diagnosis of colorectal cancer and its prognostic value. Int J Clin Exp Pathol 11:1399–1406
Zhang X, Xu Y, Qian Z, Zheng W, Wu Q, Chen Y, Zhu G, Liu Y, Bian Z, Xu W et al (2018g) CircRNA_104075 stimulates YAP-dependent tumorigenesis through the regulation of HNF4a and may serve as a diagnostic marker in hepatocellular carcinoma. Cell Death Dis 9:1091
Zhang X, Zhou H, Jing W, Luo P, Qiu S, Liu X, Zhu M, Liang C, Yu M, Tu J (2018h) The circular RNA hsa_circ_0001445 regulates the proliferation and migration of hepatocellular carcinoma and may serve as a diagnostic biomarker. Dis Markers 2018:3073467
Zhang Y, Zhang H, An M, Zhao B, Ding H, Zhang Z, He Y, Shang H, Han X (2018i) Crosstalk in competing endogenous RNA networks reveals new circular RNAs involved in the pathogenesis of early HIV infection. J Transl Med 16:332
Zhang Z, Yang T, Xiao J (2018j) Circular RNAs: promising biomarkers for human diseases. EBioMedicine 34:267–274
Zhang J, Li J, Saucier JB, Feng Y, Jiang Y, Sinson J, McCombs AK, Schmitt ES, Peacock S, Chen S et al (2019a) Non-invasive prenatal sequencing for multiple Mendelian monogenic disorders using circulating cell-free fetal DNA. Nat Med 25:1–9
Zhang Y, Chen Y, Yao H, Lie Z, Chen G, Tan H, Zhou Y (2019b) Elevated serum circ_0068481 levels as a potential diagnostic and prognostic indicator in idiopathic pulmonary arterial hypertension. Pulm Circ 9:2045894019888416
Zhang P-F, Gao C, Huang X-Y, Lu J-C, Guo X-J, Shi G-M, Cai J-B, Ke A-W (2020a) Cancer cell-derived exosomal circUHRF1 induces natural killer cell exhaustion and may cause resistance to anti-PD1 therapy in hepatocellular carcinoma. Mol Cancer 19:110
Zhang S, Song G, Yuan J, Qiao S, Xu S, Si Z, Yang Y, Xu X, Wang A (2020b) Circular RNA circ_0003204 inhibits proliferation, migration and tube formation of endothelial cell in atherosclerosis via miR-370-3p/TGFbetaR2/phosph-SMAD3 axis. J Biomed Sci 27:11
Zhao Z, Li X, Gao C, Jian D, Hao P, Rao L, Li M (2017a) Peripheral blood circular RNA hsa_circ_0124644 can be used as a diagnostic biomarker of coronary artery disease. Sci Rep 7:39918
Zhao Z, Li X, Jian D, Hao P, Rao L, Li M (2017b) Hsa_circ_0054633 in peripheral blood can be used as a diagnostic biomarker of pre-diabetes and type 2 diabetes mellitus. Acta Diabetol 54:237–245
Zhao K, Zhao Q, Guo Z, Chen Z, Hu Y, Su J, Chen L, He Z, Cai X, Chen M et al (2018a) Hsa_circ_0001275: a potential novel diagnostic biomarker for postmenopausal osteoporosis. Cell Physiol Biochem 46:2508–2516
Zhao Q, Chen S, Li T, Xiao B, Zhang X (2018b) Clinical values of circular RNA 0000181 in the screening of gastric cancer. J Clin Lab Anal 32:e22333
Zhao T, Zheng Y, Hao D, Jin X, Luo Q, Guo Y, Li D, Xi W, Xu Y, Chen Y et al (2019) Blood circRNAs as biomarkers for the diagnosis of community-acquired pneumonia. J Cell Biochem 120:16483–16494
Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, Luo Y, Lyu D, Li Y, Shi G et al (2016) Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun 7:11215
Zheng S, Gu T, Bao X, Sun J, Zhao J, Zhang T, Zhang L (2019) Circular RNA hsa_circ_0014243 may serve as a diagnostic biomarker for essential hypertension. Exp Ther Med 17:1728–1736
Zheng S, He X, Sun J, Li Q, Zhang T, Zhang L (2020) The up-regulated hsa-circRNA9102-5 may be a risk factor for essential hypertension. J Clin Lab Anal. https://doi.org/10.1002/jcla.23339
Zhou T, Xie X, Li M, Shi J, Zhou JJ, Knox KS, Wang T, Chen Q, Gu W (2018a) Rat BodyMap transcriptomes reveal unique circular RNA features across tissue types and developmental stages. RNA 24:1443–1456
Zhou X, Liu HY, Wang WY, Zhao H, Wang T (2018b) Hsa_circ_0102533 serves as a blood-based biomarker for non-small-cell lung cancer diagnosis and regulates apoptosis in vitro. Int J Clin Exp Pathol 11:4395–4404
Zhou W, Sailani RM, Contrepois K, Zhou Y, Ahadi S, Leopold SR, Zhang MJ, Rao V, Avina M, Mishra T et al (2019a) Longitudinal multi-omics of host–microbe dynamics in prediabetes. Nature 569:663–671
Zhou Z, Sun B, Huang S, Zhao L (2019b) Roles of circular RNAs in immune regulation and autoimmune diseases. Cell Death Dis 10:503
Zhu X, Wang X, Wei S, Chen Y, Chen Y, Fan X, Han S, Wu G (2017) hsa_circ_0013958: a circular RNA and potential novel biomarker for lung adenocarcinoma. FEBS J 284:2170–2182
Zhu K, Niu L, Wang J, Wang Y, Zhou J, Wang F, Cheng Y, Zhang Q, Li H (2019a) Circular RNA hsa_circ_0000885 levels are increased in tissue and serum samples from patients with osteosarcoma. Med Sci Monit 25:1499–1505
Zhu X, Ding J, Wang B, Wang J, Xu M (2019b) Circular RNA DLGAP4 is down-regulated and negatively correlates with severity, inflammatory cytokine expression and pro-inflammatory gene miR-143 expression in acute ischemic stroke patients. Int J Clin Exp Pathol 12:941–948
Zhuang Z-G, Zhang J-A, Luo H-L, Liu G-B, Lu Y-B, Ge N-H, Zheng B-Y, Li RX, Chen C, Wang X et al (2017) The circular RNA of peripheral blood mononuclear cells: Hsa_circ_0005836 as a new diagnostic biomarker and therapeutic target of active pulmonary tuberculosis. Mol Immunol 90:264–272
Zuo L, Li C, Zu J, Yao H, Yan F (2020) Circular RNA FUNDC1 improves prediction of stroke associated infection in acute ischemic stroke patients with high risk. Biosci Rep. https://doi.org/10.1042/BSR20200902
ACKNOWLEDGEMENTS
We thank Ms. Aiping Zhang for editing the figures. This work was funded by Grants from National Key R&D Program of China (2018YFC1314900, 2018YFC1314902), National Natural Science Foundation of China (61571109), and the Fundamental Research Funds for the Central Universities (2242017K3DN04).
ABBREVIATIONS
AUC, area under the receiver operating characteristic curve; CDK2, cyclin-dependent kinase 2; cfDNA, cell-free DNA; cfRNA, cell-free RNA; circ-DB, circ-deubiquitination; circRNA, circular RNA; ciRNA, circular intronic RNA; COPD, chronic obstructive pulmonary disease; CTC, circulating tumor cells; ecircRNA, exonic circular RNA; EGFR, epidermal growth factor receptor; EHI, early human immunodeficiency virus infection; EIcircRNA, exon-intron circular RNA; EV, extracellular vesicle; exLR, extracellular vesicle long RNA; f-circRNA, fusion circRNA; GC, gastric cancer; HCC, hepatocellular carcinoma; HIV, human immunodeficiency virus; ICS, intronic complement sequence; IRES, internal ribosome entry site; LPS, lipopolysaccharide; m6A, N6-methyladenosine; MBL, muscleblind; miRNA, microRNA; MRE, microRNA response element; NSCLC, non-small cell lung cancer; p21, cyclin-dependent kinase inhibitor 1; PBMC, peripheral blood mononuclear cell; PKR, protein kinase; Pol II, polymerase II; RA, rheumatoid arthritis; RBC, red blood cell; RBP, RNA binding protein; RNA-seq, RNA-sequencing; RT-ddPCR, reverse transcription droplet digital PCR; RT-qPCR, reverse transcription and quantitative PCR; SCLC, small cell lung cancer; SLE, systemic lupus erythematosus; ss, splice site; T2DM, type 2 diabetes mellitus; TB, tuberculosis.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Guoxia Wen, Tong Zhou and Wanjun Gu declare that they have no conflict of interest. This article does not contain any studies with human or animal subjects performed by the any of the authors.
OPEN ACCESS
This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (https://creativecommons.org/ licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wen, G., Zhou, T. & Gu, W. The potential of using blood circular RNA as liquid biopsy biomarker for human diseases. Protein Cell 12, 911–946 (2021). https://doi.org/10.1007/s13238-020-00799-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13238-020-00799-3