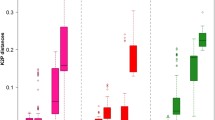
Abstract
Fungi of Nectriaceae are economically important and of high species diversity. For the purpose of accurate and rapid species identification, ITS, 28S rDNA, β-tubulin gene and EF-1α gene were selected as the candidate DNA barcode markers to investigate their feasibility in identification of 28 well-circumscribed species belonging to 9 genera of the nectriaceous fungi. A total of 216 sequences of the candidate genes were analyzed. Intra- and inter-specific variations and success rate of PCR amplification and sequencing were considered as important criteria to estimate the candidate genes. The partial β-tubulin gene met the requirements for an ideal DNA barcode and functions well for correct species delimitation. No overlapping between the intra- and inter-specific pairwise distances was found. The smallest inter-specific distance of β-tubulin gene was 3.45%, while the largest intra-specific distance was 2.77%; which appeared to possess the appropriate intra- and inter-specific variations. Twenty-eight clusters were recognized in accordance with the 28 morphological species tested. In addition, it had a high PCR and sequencing success rate. As to the other candidates, EF-1α gene showed fairly good sequence variations among species, but the PCR and sequencing success rate reached only 75.3%. ITS had a high PCR and sequencing success rate (93.5%) and recognizes 92.9% of the total number of species, nevertheless, overlapping occurred between the intra- and inter-specific distances, which may lead to incorrect species identification. 28S rDNA is most conservative compared with any other candidate markers and able to recognize merely 60.7% of the total species. We propose β-tubulin gene as the possible barcode for the nectriaceous fungi.


Similar content being viewed by others
References
Armstrong KF, Ball SL (2005) DNA barcodes for biosecurity: invasive species identification. Phil Trans R Soc B 360:1813–1823
Ball SL, Armstrong KF (2006) DNA barcodes for insect pest identification: a test case with tussock moths (Lepidoptera: Lymantriidae). Can J For Res 36:337–350
Booth C (1971) The genus Fusarium. Commonwealth Mycological Institute, Kew, Surrey
Carbone I, Kohn LM (1999) A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91:553–556
Druzhinina IS, Kopchinskiy AG, Komoń M, Bissett J, Szakacs G, Kubicek CP (2005) An oligonucleotide barcode for species identification in Trichoderma and Hypocrea. FungGenet Biol 42:813–828
Feau N, Vialle A, Allaire M, Tanguay P, Joly DL, Frey P, Callan BE, Hamelin RC (2009) Fungal pathogen (mis-) identifications: A case study with DNA barcodes on Melampsora rusts of aspen and white poplar. Mycol Res 113:713–724
Fell JW, Boekhout T, Fonseca A, Scorzetti G, Statzell-Tallman A (2000) Biodiversity and systematics of basidiomycetous yeasts as determined by large-subunit rDNA D1/D2 domain sequence analysis. Int J Syst Evol Microbiol 50:1351–1371
Frøslev TG, Jeppesen TS, Læssøe T, Kjøller R (2007) Molecular phylogenetics and delimitation of species in Cortinarius section Calochroi (Basidiomycota, Agaricales) in Europe. Mol Phylogenet Evol 44:217–227
Geiser DM, Jiménez-Gasco M, Kang S, Makalowska I, Veeraraghavan N, Ward TJ, Zhang N, Kuldau GA, O’Donnell K (2004) FUSARIUM-ID v.1.0: A DNA sequence database for identifying Fusarium. Eur J Plant Pathol 110:473–479
Geiser DM, Klich MA, Frisvad JC, Peterson SW, Varga J, Samson RA (2007) The current status of species recognition and identification in Aspergillus. Stud Mycol 59:1–10
Glass NL, Donaldson GC (1995) Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl Environ Microb 61:1323–1330
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Hebert PD, Cywinska A, Ball SL, de Waard JR (2003a) Biological identifications through DNA barcodes. Proc R Soc Lond 270:313–321
Hebert PD, Ratnasingham S, de Waard JR (2003b) Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proc R Soc Lond B (Suppl) 270:S96–S99
Hebert PD, Stoeckle MY, Zemlak TS, Francis CM (2004) Identification of Brids through DNA Barcodes. PLoS Biol 2:e312
Hebert PD, de Waard JR, Landry J-F (2009) DNA barcodes for 1/1000 of the animal kingdom. Biol Lett. doi:10.1098/rsbl.2009.0848
Hollingsworth PM, Forrest LL, Spouge JL et al (2009) A DNA barcode for land plants. Proc Natl Acad Sci USA 106:12794–12797
Johnson J, Vilgalys R (1998) Phylogenetic systematics of Lepiota sensu lacto based on nuclear large subunit rDNA evidence. Mycologia 90:971–979
Kirk PM, Cannon PF, Minter DW, Stalpers JA (eds) (2008) Dictionary of the Fungi (10th ed). CAB International. Wallingford, UK
Kurtzman CP, Robnett CJ (1998) Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26 S) ribosomal DNA partial sequences. Antonie van Leeuwenhoek 73:331–371
Li J, Liu SC, Niu SB, Zhuang WY, Che YS (2009) Pyrrolidinones from the ascomycete fungus Albonectria rigidiuscula. J Nat Prod 73:2184–2187
Luo J, Zhuang WY (2010) Three new species of Neonectria (Nectriaceae, Hypocreales) with notes on their phylogenetic positions. Mycologia 102:142–152
Martens M, Dawyndt P, Coopman R, Gillis M, De Vos P, Willems A (2008) Advantages of multilocus sequence analysis for taxonomic studies: a case study using 10 housekeeping genes in the genus Ensifer (including former Sinorhizobium). Int J Syst Evol Microbiol 58:200–214
Meier R, Shiyang K, Vaidya G, Ng PK (2006) DNA barcoding and taxonomy in Diptera: A tale of high intraspecific variability and low identification success. Syst Biol 55:715–728
Meyer CP, Paulay G (2005) DNA barcoding: Error rates based on comprehensive sampling. PLoS Biol 3:e422
Ninet B, Jan I, Bontems O, Léchenne B, Jousson O, Panizzon R, Lew D, Monod M (2003) Identification of dermatophyte species by 28 S ribosomal DNA sequencing with a commercial kit. J Clin Microbiol 41:826–830
O’Donnell K, Cigelnik E (1997) Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Mol Phylogenet Evol 7:103–116
Rehner SA, Samuels GJ (1994) Taxonomy and phylogeny of Gliocladium analyzed from nuclear large subunit ribosomal DNA sequences. Mycol Res 98:625–634
Rehner SA, Samuels GJ (1995) Molecular systematics of the Hypocreales: A teleomorph gene phylogeny fad the status of their anamorph. Can J Bot 73(suppl I):S816–S823
Rehner SA, Buckley E (2005) A Beauveria phylogeny inferred from nuclear ITS and EF1-α sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 97:84–98
Roe AD, Rice AV, Bromilow SE, Cooke JEK, Sperling FAH (2010) Multilocus species identification and fungal DNA barcoding: insights from blue stain fungal symbionts of the mountain pine beetle. Mol Ecol Res doi:. doi:10.1111/j.1755-0998.2010.02844.x
Rossman AY (1996) Morphological and molecular perspectives on systematics of the Hpocreales. Mycologia 88:1–19
Rossman AY, Samuels GJ, Rogerson CT, Lowen R (1999) Genera of Bionectriaceae, Hypocreaceae and Nectriaceae (Hypocreales, Ascomycetes). Stud Mycol 42:1–248
Schroers H-J, Geldenhuis MM, Wingfield MJ, Schoeman MH, Yen Y-F, Shen W-C, Wingfield BD (2005) Classification of the guava wilt fungus Myxosporium psidii, the palm pathogen Gliocladium vermoesenii and the persimmon wilt fungus Acremonium diospyri in Nalanthamala. Mycologia 97:375–395
Schwarz P, Bretagne S, Gantier J-C, Garcia-Hermoso D, Lortholary O, Dromer F, Dannaoui E (2006) Molecular identification of zygomycetes from culture and experimentally infected tissues. J Clin Microbiol 44:340–349
Seifert KA (2009) Progress towards DNA barcoding of fungi. Mol Ecol Resour 9(Suppl 1):83–89
Slabbinck B, Dawyndt P, Martens M, De Vos P, De Baets B (2008) TaxonGap: a visualisation tool for intra- and inter-species variation among individual biomarkers. Bioinformatics 24:866–867
Stoeckle MY, Hebert PD (2008) Barcode of life. Sci Am 299(82–86):88
Taberlet P, Coissac E, Pompanon F, Gielly L, Miquel C, Valentini A, Vermat T, Corthier G, Brochmann C, Willerslev E (2007) Power and limitations of the chloroplast trnL (UAA) intron for plant DNA barcoding. Nucleic Acids Res 35:e14
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignments through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680
Valentini A, Pompanon F, Taberlet P (2009) DNA barcoding for ecologists. Trends Ecol Evol 24:110–117
Vilgalys R, Hester M (1990) Rapid genetic identification and mapping enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol 172:4238–4246
Wang L, Zhuang WY (2004) Designing primer sets for amplification of partial calmodulin genes from penicillia. Mycosystema 23:466–473
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR Protocols: A Guide to Methods and Applications. Academic, New York, pp 315–322
Zhang XM, Zhuang WY (2006) Phylogeny of some genera in Nectriaceae (Ascomycetes, Hypocreales) inferred from rDNA 28 S partial sequences. Mycosystema 25:15–22
Acknowledgements
The authors express their deep thanks to Prof. R. P. Korf for corrections of the language, and Dr. R. Meier for providing a useful reference. This project was supported by the State 863 Project (no. 2008AA02Z312) and Special Project for Fundamental Research (no. 2006FY120100) from the Ministry of Science and Technology of China, and the National Natural Science Foundation of China (no. 31070015) to WYZ.
Author information
Authors and Affiliations
Corresponding author
Additional information
Peng Zhao and Jing Luo contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Suppl. Fig. 1
Neighbor-joining tree based on ITS sequences from 28 species of Nectriaceae with Bionectria wenpingii as outgroup. TreeBase S10405 (GIF 163 kb)
Suppl. Fig. 2
Neighbor-joining tree based on 28 S rDNA sequences from 28 species of Nectriaceae with Bionectria wenpingii as outgroup. TreeBase S10406 (GIF 190 kb)
Suppl. Fig. 3
Neighbor-joining tree based on β-tubulin gene sequences from 28 species of Nectriaceae with Bionectria wenpingii as outgroup. TreeBase S10407 (GIF 173 kb)
Suppl. Fig. 4
Neighbor-joining tree based on EF-1α gene sequences from 28 species of Nectriaceae with Bionectria wenpingii as outgroup. TreeBase S10408 (GIF 184 kb)
Rights and permissions
About this article
Cite this article
Zhao, P., Luo, J. & Zhuang, WY. Practice towards DNA barcoding of the nectriaceous fungi. Fungal Diversity 46, 183–191 (2011). https://doi.org/10.1007/s13225-010-0064-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13225-010-0064-y