Abstract
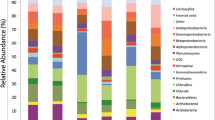
Pseudomonas community structures were investigated by analyzing 16S rRNA clone libraries derived from fertilized and unfertilized soil plots under corn–alfalfa rotation in a long-term experiment. Amplified 16S rRNA fragments derived by polymerase chain reaction (PCR) were cloned and sequenced. A total of 729 clone sequences were analyzed, of which 51 were possible chimeras and discarded. The remaining clone sequences (678) belonged to γ-proteobacteria with 61.8 % (419) classified to the genus Pseudomonas. Unclassified Gammaproteobacteria accounted for 23.4 % of total clones sequences. Rarefaction analyses showed a more diverse community structure of both Gammaproteobacteria and Pseudomonas in unfertilized than fertilized field soils irrespective of plant types under cultivation. Bacterial or Pseudomonas community structures differed significantly between fertilized and unfertilized soil plots. Clone sequences that are affiliated to Pseudomonas putida and P. oryzihabitans were more prominent in libraries from fertilized plots, while those that clustered with Pseudomonas frederiksbergensis were more often retrieved from unfertilized soil plots. A strong influence of fertilizer applications on community structure was supported by principal component analysis. We conclude that long-term use of mineral fertilizers could influence Pseudomonas community structure.





Similar content being viewed by others
References
Amann RI, Ludwig W, Schleifer KH (1995) Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev 59:143–169
Anzai Y, Kudo Y, Oyaizu H (1997) The phylogeny of the genera Chryseomonas, Flavimonas, and Pseudomonas supports synonymy of these three genera. Int J Syst Bacteriol 47:249–251
Ashelford KE, Chuzhanova NA, Fry JC, Jones AJ, Weightman AJ (2006) New screening software shows that most recent large 16 S rRNA gene clone libraries contain chimeras. Appl Environ Microbiol 72:5734–5741
Baati H, Guermazi S, Gharsallah N, Sghir A, Ammar E (2010) Microbial community of salt crystals processed from Mediterranean seawater based on 16 S rRNA analysis. Can J Microbiol 56:44–51
Bolwerk A, Lagopodi AL, Wijfjes AH, Lamers GE, Chin AWTF, Lugtenberg BJ, Bloemberg GV (2003) Interactions in the tomato rhizosphere of two Pseudomonas biocontrol strains with the phytopathogenic fungus Fusarium oxysporum f. sp. radicis-lycopersici. Mol Plant Microbe Interact 16:983–993
Buckley DH, Schmidt TM (2003) Diversity and dynamics of microbial communities in soils from agro-ecosystems. Environ Microbiol 5:441–452
Carter JP, Richardson DJ, Spiro S (1995) Isolation and characterisation of a strain of Pseudomonas putida that can express a periplasmic nitrate reductase. Arch Microbiol 163:159–166
Clarke KR (1993) Non-parametric multivariate analysis of changes in community structure. Aust J Ecol 18:117–143
Clausen GB, Larsen L, Johnsen K, Radnoti de Lipthay J, Aamand J (2002) Quantification of the atrazine-degrading Pseudomonas sp. strain ADP in aquifer sediment by quantitative competitive polymerase chain reaction. FEMS Microbiol Ecol 41:221–229
Colwell RR (2009) Viable but not cultivable bacteria. In: Epstein SS (ed) Uncultivated microorganisms. Springer, Berlin, p 220
Costa R, Gomes NC, Krogerrecklenfort E, Opelt K, Berg G, Smalla K (2007) Pseudomonas community structure and antagonistic potential in the rhizosphere: insights gained by combining phylogenetic and functional gene-based analyses. Environ Microbiol 9:2260–2273
Cruz-Martinez K, Suttle KB, Brodie EL, Power ME, Andersen GL, Banfield JF (2009) Despite strong seasonal responses, soil microbial consortia are more resilient to long-term changes in rainfall than overlying grassland. Inter Soc Microbial Ecol J 3:738–744
Drury CF, Tan CS (1995) Long-term (35 years) effects of fertilizstion, rotation and weather on corn yields. Can J Plant Sci 75:355–362
Drury CF, Yang XM, Reynolds WD, Tan CS (2004) Influence of crop rotation and aggregate size on carbon dioxide and nitrous oxide emissions. Soil and Tillage Res 79:87–100
Duineveld BM, Kowalchuk GA, Keijzer A, van Elsas JD, van Veen JA (2001) Analysis of bacterial communities in the rhizosphere of chrysanthemum via denaturing gradient gel electrophoresis of PCR-amplified 16 S rRNA as well as DNA fragments coding for 16 S rRNA. Appl Environ Microbiol 67:172–178
Dunbar J, Takala S, Barns SM, Davis JA, Kuske CR (1999) Levels of bacterial community diversity in four arid soils compared by cultivation and 16 S rRNA gene cloning. Appl Environ Microbiol 65:1662–1669
Dunn AK, Stabb EV (2005) Culture-independent characterization of the microbiota of the ant lion Myrmeleon mobilis (Neuroptera: Myrmeleontidae). Appl Environ Microbiol 71:8784–8794
Endert E, Ritchie DF (1984) Detection of pathogenicity, measurement of virulence, and determination of strain variation in Pseudomonas syringae pv. syringae. Plant Dis 68:677–680
Ferrante P, Scortichini S (2010) Molecular and phenotypic features of Pseudomonas syringae pv. actinidiae isolated during recet epidemics of bacterial canker on yellow kiwifruit (Actinidia chinensis) in central Italy. Plant Pathol 59:954–962
Garbeva P, van Veen JA, van Elsas JD (2004) Microbial diversity in soil: selection microbial populations by plant and soil type and implications for disease suppressiveness. Annu Rev Phytopathol 42:243–270
Garrity GM, Bell JA, Lilburn TG (2004) Taxomonic outline of the prakaryotes, Bergey’s manual of systematic bacteriology, 2nd edn. Springer, New York
Hayward AC, Fegan N, Fegan M, Stirling GR (2010) Stenotrophomonas and Lysobacter: ubiquitous plant-associated gamma-proteobacteria of developing significance in applied microbiology. J Appl Microbiol 108:756–770
Ibekwe AM, Papiernik SK, Gan J, Yates SR, Yang CH, Crowley DE (2001) Impact of fumigants on soil microbial communities. Appl Environ Microbiol 67:3245–3257
Kim M, Jeong SY, Yoon SJ, Cho SJ, Kim YH, Kim MJ, Ryu EY, Lee SJ (2008) Aerobic denitrification of Pseudomonas putida AD-21 at different C/N ratios. J Biosci Bioeng 106:498–502
Lemanceau P, Corberand T, Gardan L, Latour X, Laguerre G, Boeufgras J, Alabouvette C (1995) Effect of Two Plant Species, Flax (Linum usitatissinum L.) and Tomato (Lycopersicon esculentum Mill.), on the diversity of soilborne populations of fluorescent pseudomonads. Appl Environ Microbiol 61:1004–1012
Lin W-M (1999) Denitrification by Pseudomonas putida with a fluidized-bed column. Georgia State University, Atlanta
Lin YT, Huang YJ, Tang SL, Whitman WB, Coleman DC, Chiu CY (2010) Bacterial community diversity in undisturbed perhumid montane forest soils in Taiwan. Microbial Ecol 59:369–378
Lozupone C, Knight R (2005) UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol 71:8228–8235
Ludwig W, Bauer SH, Bauer M, Held I, Kirchhof G, Schulze R, Huber I, Spring S, Hartmann A, Schleifer KH (1997) Detection and in situ identification of representatives of a widely distributed new bacterial phylum. FEMS Microbiol Lett 153:181–190
Mazurier S, Corberand T, Lemanceau P, Raaijmakers JM (2009) Phenazine antibiotics produced by fluorescent pseudomonads contribute to natural soil suppressiveness to Fusarium wilt. Inter Soc Microbial Ecol J 3:977–991
McCaig AE, Glover LA, Prosser JI (1999) Molecular analysis of bacterial community structure and diversity in unimproved and improved upland grass pastures. Appl Environ Microbiol 65:1721–1730
Milling A, Smalla K, Maidl FX, Schloter M, Munch JC (2004) Effects of transgenic potatoes with an altered starch composition on the diversity of soil and rhizosphere bacteria and fungi. Plant Soil 266:23–39
Mulet M, Lalucat J, García-Valdés E (2010) DNA sequence-based analysis of the Pseudomonas species. Environ Microbiol 12:1513–1530
Muyzer G, de Waal EC, Uitterlinden AG (1993) Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16 S rRNA. Appl Environ Microbiol 59:695–700
O'Donnell AG, Gorres HE (1999) 16 S rDNA methods in soil microbiology. Curr Opin Biotechnol 10:225–229
O'Donnell AG, Seasman M, Macrae A, Waite I, Davies JT (2001) Plants and fertilisers as drivers of change in microbial community structure and function in soils. Plant Soil 232:135–145
Palleroni N (2005) Genus I. Pseudomonas Migula. In: Garrity GM, Brenner DJ, Krieg NR, Staley JT (eds) Part B: the Gammaproteobacteria, Bergey’s manual of systematic bacteriology, 2nd edn. Springer, New York
Park SH, Lee K, Chae JC, Kim CK (2004) Construction of transformant reporters carrying fused genes using pcbC promoter of Pseudomonas sp DJ-12 for detection of aromatic pollutants. Environ Monit Assess 92:241–251
Patten CL, Glick BR (2002) Role of Pseudomonas putida indoleacetic acid in development of the host plant root system. Appl Environ Microbiol 68:3795–3801
Peacock AD, Mullen MD, Ringelberg DB, Tyler DD, Hedrick DB, Gale PM, White DC (2001) Soil microbial community responses to dairy manure or ammonium nitrate applications. Soil Biol Biochem 33:1011–1019
Posada D (2009) Selection of models of DNA evolution with jModelTest. Methods Mol Biol 537:93–112
Postma J, Schilder MT, Bloem J, Van Leeuwen-Haagsma WK (2008) Soil suppressiveness and functional diversity of the soil microflora in organic farming systems. Soil Biol Biochem 40:2394–2406
Raaijmakers JM, Weller DM (2001) Exploiting genotypic diversity of 2,4-diacetylphloroglucinol-producing Pseudomonas spp.: characterization of superior root-colonizing P. fluorescens strain Q8r1-96. Appl Environ Microbiol 67:2545–2554
Sarkar SF, Guttman DS (2004) The evolution of the core genome of Pseudomonas syringae, a highly clonal, endemic plant pathogen. Appl Environ Microbiol 70:1999–2012
Schloss PD, Handelsman J (2005) Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl Environ Microbiol 71:1501–1506
Schloss PD, Larget BR, Handelsman J (2004) Integration of microbial ecology and statistics: a test to compare gene libraries. Appl Environ Microbiol 70:5485–5492
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541
Singleton DR, Furlong MA, Rathbun SL, Whitman WB (2001) Quantitative comparisons of 16 S rRNA gene sequence libraries from environmental samples. Appl Environ Microbiol 67:4374–4376
Smit E, Leeflang P, Gommans S, van den Broek J, van Mil S, Wernars K (2001) Diversity and seasonal fluctuations of the dominant members of the bacterial soil community in a wheat field as determined by cultivation and molecular methods. Appl Environ Microbiol 67:2284–2291
Sun HY, Deng SP, Raun WR (2004) Bacterial community structure and diversity in a century-old manure-treated agroecosystem. Appl Environ Microbiol 70:5868–5874
Tambong JT, Höfte M (2001) Phenazines are involved in biocontrol of Pythium myriotylum on Cocoyam by Pseudomonas aeruginosa PNA1. Eur J Plant Pathol 107:511–521
Tambong JT, de Cock AW, Tinker NA, Levesque CA (2006) Oligonucleotide array for identification and detection of Pythium species. Appl Environ Microbiol 72:2691–2706
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Torsvik V, Salte K, Sorheim R, Goksoyr J (1990) Comparison of phenotypic diversity and DNA heterogeneity in a population of soil bacteria. Appl Environ Microbiol 56:776–781
Torsvik V, Sørheim R, Goksøyr J (1996) Total bacterial diversity in soil and sediment communities-a review. Ind Microbiol 17:170–178
van Elsas JD, Garbeva P, Salles J (2002) Effects of agronomical measures on the microbial diversity of soils as related to the suppression of soil-borne plant pathogens. Biodegradation 13:29–40
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73:5261–5267
Widmer F, Seidler RJ, Gillevet PM, Watrud LS, Di Giovanni GD (1998) A highly selective PCR protocol for detecting 16 S rRNA genes of the genus Pseudomonas (sensu stricto) in environmental samples. Appl Environ Microbiol 64:2545–2553
Acknowledgement
Funding for this work was provided by Agriculture and Agri-Food Canada through project # 152. We are indebted to C. Drury, for managing the long-term experimental plots at Woodslee, Ontario, Canada. We are also grateful to T. Barasubiye for extraction of total soil DNA, and Rafik Assabgui for technical assistance in DNA sequencing.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Tambong, J.T., Xu, R. Culture-independent analysis of Pseudomonas community structures in fertilized and unfertilized agricultural soils. Ann Microbiol 63, 323–333 (2013). https://doi.org/10.1007/s13213-012-0477-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13213-012-0477-9