Abstract
The pandemic COVID-19 was caused by a novel Coronavirus-2 (SARS-CoV-2) that infects humans through the binding of glycosylated SARS-CoV-2 spike 2 protein to the glycosylated ACE2 receptor. The spike 2 protein recognizes the N-terminal helices of the glycosylated metalloprotease domain in the human ACE2 receptor. To understand the susceptibility of animals for infection and transmission, we did sequence and structure-based molecular interaction analysis of 16 ACE2 receptors from different mammalian species with SARS-CoV-2 spike 2 receptor binding domain. Our comprehensive structure analysis revealed that the natural substitution of amino acid residues Gln24, His34, Phe40, Leu79 and Met82 in the N-terminal α1 and α2 helices of the ACE2 receptor results in loss of crucial network of hydrogen-bonded and hydrophobic interactions with receptor binding domain of SARS-CoV-2 spike protein. Another striking observation is the absence of N-glycosylation site Asn103 in all mammals and many species, lack more than one N-linked glycosylation site in the ACE2 receptor. Based on the loss of crucial interactions and the absence of N-linked glycosylation sites we categorized Felis catus, Equus caballus, Panthera tigris altaica, as highly susceptible while Oryctolagus cuniculus, Bos Tauras, Ovis aries and Capra hircus as moderately susceptible species for infection. Similarly, the E. asinus, Bubalus bubalis, Canis lupus familiaris, Ailuropoda melaleuca and Camelus dromedarius are categorized as low susceptible with Loxodonta Africana, Mus musculus, Sus scrofa and Rattus rattus as least susceptible species for SARS-CoV-2 infection.
Similar content being viewed by others
Introduction
COVID-19 is one of the most dreadful pandemic diseases of the 21st century responsible for most deaths worldwide. The first human COVID-19 case was reported from Wuhan city, Hubei state of China, in December 2019 and expanded globally as a new health pandemic (Zhao et al. 2020b) (Li et al. 2020). As reported by World Health Organization till 12th December 2020, 69.5 million cases are confirmed with infection globally, and the total death is 1.5 million (World Health Organization). Belonging to the realm of Ribovaria, family Coronaviridae, suborder Coronovirineae and order Nidovirales (Contini et al. 2020)(Kannan et al. 2020)(Hasöksüz et al. 2020), the Severe Acute Respiratory Syndrome related coronaviruses are mostly zoonotic (Chen et al. 2020). The phylogenetic analysis showed these as four distinct groups (Bhowmik et al. 2020). Hypothesized to have originated in bats with unknown intermediate hosts, this could be the result of a “spillover event”(WHO) (Banerjee et al. 2019). Earlier studies described bats and pangolins as intermediate hosts (Zhang et al. 2020a); subsequent literature failed to reveal any such connections (Zhang et al. 2020b). Many zoonotic CoVs are sustaining in nature, constantly mutating and evolving to give rise to a new kind of infectious form (Ye et al. 2020). The person infected with this virus develops various symptoms such as fever, body ache, cough, pneumonia etc. (Singhal 2020). Some of these coronaviruses can transmit between animals and humans (Cevik et al. 2020) mainly through direct physical contacts, droplet transmission or oral transmission and prevalent in all age groups. Coronaviruses have four genera: α, β, γ, δ and the novel coronavirus (SARS-CoV-2) (Gabutti et al. 2020) belongs to β type Coronavirus. The Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a positive-sense single-stranded RNA virus that contains four main structural proteins (Mousavizadeh and Ghasemi 2020) the spike glycoprotein (S), non-structural proteins (Nsps), membrane glycoprotein (M), and several accessory (Jiang et al. 2020) protein with a diameter of 80–120 nm. It consists of a large genome of 28–32 kb. To enter the host cell, the SARS-CoV-2 virus utilizes trimeric glycosylated spike protein to bind the glycosylated ACE2 receptor. The spike protein uses receptor-binding subunit S1 to attach to the ACE2 receptor and the subunit S2 for membrane fusion into the host cell (Yan et al. 2020). The trimeric SARS-CoV-2 Spike protein is shielded by 66 possible N-linked glycosylation sites thereby escape host immune invasion and form a stable complex with the ACE2 receptor. Similarly, the host cell ACE2 receptor is also glycosylated with the N-terminal extracellular domain containing seven N-linked and three O-linked glycosylation sites. The alteration in glycosylation of the ACE2 receptor reduces the affinity for SARS-CoV-2 Spike protein (Donoghue et al. 2000; Tipnis et al. 2000; Li et al. 2005; Schwarz and Aebi 2011a). By glycomic and glycoproteomic approach, Wells group analyzed the purified complex of SARS-CoV-2 spike protein and the human ACE2 receptor shown that the natural variant in the ACE2 receptor glycosylation influences the SARS-CoV-2 Spike protein binding (Zhao et al. 2020a). For a mammalian species to be highly susceptible to infection the ACE2 receptor has to be properly glycosylated. The sugar moieties at the glycosylation site act as a ligand for cell surface receptors to mediate cell attachment or induce a signal transduction pathway (Ohtsubo and Marth 2006). The studies have shown that the glycosylation pattern influences viral entry into the host and pathogenesis (Han et al. 2007a; Almendros and Gascoigne 2020; Shi et al. 2020).To the glycosylated receptor, the SARS-CoV-2 attaches to the concave binding surface of the receptor with (Schwarz and Aebi 2011b) spike receptor-binding domain by a hinge-like motion (Towler et al. 2004; Han et al. 2007b; Schwarz and Aebi 2011b; Shang et al. 2020). Interestingly, the animal with direct contact with humans has minimal susceptibility to SARS-CoV-2 infection (Baloch et al. 2020). A study by shi et al. based on the clinical samples has shown that cats and ferrets are more susceptible to infection than pigs, chickens, dogs and ducks dogs (Shi et al. 2020). Similarly, a recent clinical sample study conducted in pets, 9 cats and 12 dogs in a veterinary campus also indicated cats are prone to infection than dogs (Temmam et al. 2020). Till now no study has been reported based on both molecular interactions between SARS-CoV-2 spike protein and the ACE2 receptor along with glycosylation sites. In this study, we have performed a comprehensive sequence and detailed molecular interactions analysis of 16 ACE2 receptors from different mammals to understand their susceptibility for infection. The mammalian species selected in this study is based on four-category viz. common domestic animals which are in daily direct contact with humans, wild animals, animals reportedly prone to the disease, and some closely related species.
Materials and methods
Multiple sequence alignment
The sequences of the ACE2 receptor from human and 16 other common mammalian species (Fig. S1) were retrieved from the National Center for Biotechnology Information (NCBI) protein sequence database. The multiple sequence alignment was performed using the online sequence alignment tool Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/) (Sievers and Higgins 2014).
Homology modelling
The homology models of the ACE2 receptor of the 16 common mammalian species were generated using the SWISS-MODEL server (https://swissmodel.expasy.org/) (Waterhouse et al. 2018). The best model for each species was selected based on Levitt-Gerstein (LG) score (Levitt and Gerstein 1998) using the neural network-based method and ProQ server (https://proq.bioinfo.se/ProQ/ProQ.html). The chosen model of each species was refined by energy minimization using previously reported parameters (Yadav et al. 2019; Naz et al. 2020). The stereochemical quality of the 16 models was evaluated by the PROCHECK program (Laskowski et al. 1993) and ERRAT server (Colovos and Yeates 1993).
Docking of SARS-CoV-2 spike protein with ACE-2 protein
The protein–protein docking of the ACE2 receptor model with the receptor-binding domain of spike protein was performed using the HDOCK server (Yan et al. 2020b), a hybrid approach based on template-based modeling and ab initio docking. Based on the docking score, the best-docked conformation was selected for each species, and energy minimized using previously reported parameters (Yadav et al. 2019). The energy minimized docked complex of individual species was superimposed on to the crystal complex of human ACE2 and spike protein (PDB ID–6M0J and 6M18) (Brooke and Prischi 2020; Stout et al. 2020) for structural comparison and interaction analysis.
Binding free energy calculation
The binding energy of each energy minimized docked complex of SARS-CoV-2 spike protein and the respective ACE2 receptor was calculated by MM-GBSA (Molecular Mechanics-Generalized Born Surface Area) approach to estimate the relative binding affinity. Binding energy was calculated using OPLS-2005 molecular mechanics force field parameters (Shivakumar et al. 2010) and continuum (implicit) solvation models similar to the previously described approach (Beard et al. 2013).
Comparison of glycosylation pattern between human and other mammalian species
The ACE2 receptor glycosylation sites of all 16 mammalian species were predicted and analyzed using the NetNGlyc1.0 server.
Results
Multiple sequence alignment
The multiple sequence alignment of the Human ACE2 receptor with other mammalian species (Table 1) showed the highest sequence identity with Equus caballus (Horse) (86.78%) and lowest identity with the Rattus rattus (Black rat) (81.01%). Based on the structure complex of human ACE2 and spike protein (PDB ID:6M0J), the residues majorly involved in hydrogen-bonded and hydrophobic interactions with spike 2 receptor binding domain are Gln24, Asp30, Lys31, His34, Glu35, Asp38, Tyr41, Gln42, Tyr83, Lys353 and Asp355 (Fig. 1). The substitution of these residues leads to the loss of interactions which might affect affinity. Out of these 10 residues, Lys31, Glu35, Asp38, Gln42, and Tyr41 were conserved in all species. Asp30 is mostly replaced by Glu30, maintaining the residual charge integrity and Tyr41 is conserved in most species except in E. asinus and E. caballus, where Histidine replaces it. Similarly, Tyr83 and Lys353 are conserved in most species except for Mus musculus, R. rattus and Loxodonta africana, where the residues are replaced by phenylalanine and histidine, respectively. The crucial substitutions observed in the sequence alignment were Gln24 and Met82 to Leu and Thr, respectively. Besides, the absence of a glycosylation site at position Asn103 in all other mammals also plays an essential role in spike recognition of the ACE2 receptor. In human, the ACE2 receptor contains seven N-linked glycosylation sites (Asn53, Asn90, Asn103, Asn322, Asn432, Asn546 and Asn690) and three O-linked glycosylation sites (S155, S496 and S730) (Shajahan et al. 2020a). Out of seven N-linked glycosylation sites, five (Asn53, Asn90, Asn103, Asn322 and Asn546) are aligned close to the spike 2 recognition site and might have positive role in binding with spike 2 protein thereby increasing the binding affinity. Interestingly, in almost all species, the position Asn103 is replaced by Ser103 (Fig. 1), making this position exclusive for the human ACE2 receptor.
Multiple Sequence alignment of the human ACE2 receptor with 16 selected mammalian species. The key conserved residues involved in hydrogen-bonding (cyan), hydrophobic interaction (pink) with the SARS-CoV-2 spike protein in the three interacting regions are shown. The N-linked glycosylation sites in the ACE2 receptor are shown in red. The hydrophobic residues lined in the concave interacting surface (blue) and key residues at position Met82 and Phe40 of ACE2 receptor are highlighted in green and yellow color
Homology modeling
The homology model of ACE2 receptors from different species was generated using PDB 6M0J (Lan et al. 2020). The Levitt-Gerstein (LG) score of each model calculated by the ProQ server showed that the structural quality parameters are within the acceptable range (> 5.0) (Table 1). The PROCHECK analysis showed that greater than 99% of the residues fall in the allowed, additionally allowed and generously allowed regions of the Ramachandran plot (Table S1, Figs. S2, S3). The ERRAT value of all the structure models was found to be greater than 93%. The generated models have the right geometry; hence the models were further used for protein–protein docking.
Protein–protein docking
The protein–protein docking of the ACE2 receptor model with SARS-CoV-2 spike protein receptor-binding domain (RBD) was performed using the HDock server. The human ACE2 receptor and SARS-CoV-2 spike RBD domain complex (6M0J) were used as a docking reference (Lan et al. 2020). The top model (out of 10) was used for molecular interaction analysis using the Schrodinger Maestro MMGBSA method (Beard et al. 2013). The Gibbs free energy of binding for each complex was calculated and compared with human (− 128.67 kcal/mol) with 949.8 Å2 buried surface area between two proteins (Table 2). The lowest Gibbs free energy (∆G) next to human was observed for Felis catus (− 98.04 kcal/mol) followed by E. caballus (− 97.24 kcal/mol), Panthera tigris altaica (− 93.85 kcal/mol) and Oryctolagus cuniculus (-93.07 kcal/mol). Similarly, the highest ∆G was found for the complexes of Sus scrofa (− 46.31 kcal/mol) and R. rattus (− 43.21 kcal/mol) (Table 2). The free binding energy indicates that there is the loss of some crucial interactions resulting in lower affinity between SARS-CoV-2 spike protein and ACE2 receptor from other species.
Glycosylation of ACE2 receptor
Glycosylation plays a significant role in host-parasite interaction. The pattern of glycosylation contributes to the recognition of the ACE2 receptor by the SARS-CoV-2 spike protein. The glycoproteomics of the human ACE2 receptor showed the presence of seven N-linked and three O-linked glycosylation sites. By LC–MS/MS, a particular pattern of sugar moieties were reported for both N-linked and O-linked glycosylation sites. In N-linked sites, a bisecting N-acetyl Glucosamine, an N-glycan fucosylation with the sialic acid as terminal sugar moiety was observed (Shajahan et al. 2020a; Zhao et al. 2020a). While, in the crystal structures of the human ACE2 receptor (PDB ID: 6M0J and 6M18) sugar moieties were observed at the N-linked glycosylation sites Asn53, Asn90, Asn103, Asn322, Asn432, Asn546 and Asn690. Out of these seven sites, Asn53, Asn90, Asn103, Asn322 and Asn546 were observed in the close vicinity of the spike protein-interacting interface forming a pillar-like structure (Fig. 2). Based on the previously reported study (Shajahan et al. 2020a) the sugar molecules were modeled at these N-linked glycosylation sites as shown in Fig. 2b. The sugar moieties at these glycosylation sites are held by nearby supporting residues (Table 3). The Glycan chain at Asn53 is supported by Thr55, Glu57, Asn58 and Gln340. While the glycan chains at Asn90, Asn103, Asn322 and Asn546 are supported by Lys26 and Gln81, His194, Asn195 and Glu312 and Ser420, respectively. Overall, four glycosylations and supporting residual hydrogen bonding and hydrophobic interactions make the human ACE2 receptor more specific than other mammalian species. Upon sequence and structure comparison of all selected mammalian species, the Asn103 N-linked glycosylation is either absent or replaced by Ser. The NetOGlyc 4.0 server prediction also showed no possibility for glycosylation. In Canis lupus familiaris, Asn90 and Asn103 sites are absent. In Oryctolagus cuniculus, Bos taurus, Ovis aries, Capra hircus, and Bubalus bubalis Asn103 and Asn322 sites are absent. While in Loxodonta Africana, Mus musculus and R. rattus three glycosylation sites Asn90, Asn103 and Asn322 are absent. Except for Ailuropoda melanoleuca, E. asinus, E. caballus, Felis catus, and Panthera tigris altaica all other species lack more than one glycosylation site. The complete list of glycosylation sites of all the species is shown in Table 3.
The cartoon representation of human ACE2 receptor in complex with SARS-COV-2 spike-RBD domain. a The human ACE2 receptor metalloprotease domain and SARS-CoV-2 spike receptor-binding domain is shown in brown and cyan, respectively. The blue box represents a close view of the hydrogen-bonded interactions formed at three interacting regions (Region 1–3) along with four glycosylation sites are shown. Met82 residue is present in Region 4, The red box represents a closer view of the hydrogen bonding and hydrophobic network of interactions connecting Asn90 (Site 2) and Asn103 (Site 3) glycosylation sites. b Model representation of five important glycosylation sites along with the sugar moieties (Fucose, N-acetylneuraminic acid, N-acetylglucosamine, Mannose and Galactose) proposed in the study done by Varki, A., Cummings, R.D., et al. 2015
Structural comparison of ACE2 receptor and spike 2 complexes
The SARS-CoV-2 spike protein juxtaposes with human ACE2 receptors predominantly by hydrogen bonds, hydrophobic interactions, ionic interactions along with glycosylation sites. The Spike protein recognizes the concave surface of the metalloprotease domain of the ACE2 receptor. The interface can be divided into four interacting regions, majorly involves N-terminal α1, α2 helices and β3, β4 strands. The interacting region 1 is formed by the N-terminal part of α1 helix; the interacting region 2 is formed by the mid part of the α1 helix; the Interacting region 3 is formed by the end part of α1 helix and β3, β4 strands; while the interacting region 4 is formed by the end part of α2 helix (Lan et al. 2020)(Yan et al. 2020). Comparison of the sequence of ACE2 receptor from different mammals notable natural amino acid substitution was observed. A detailed structural analysis of each interacting region from all species ACE2 receptor was performed to understand the SARS-CoV-2 spike protein recognition.
SARS-CoV-2 spike protein-interacting region 1: N-terminal α1 helix of ACE2 receptor
Upon structural analysis of the complex in the SARS-CoV-2 spike protein-interacting region 1, Gln24 residue is crucial in forming a chain of three hydrogen-bonded interactions. The Gln24 (NE2) at one end holds the backbone atoms of N-terminal residues Ser19/Thr20 and in another end (Gln24 OE1) interacts with Asn487 (RBD) to position the residue to form stable hydrogen bonds with Tyr83 of α2 helix. The Tyr83 in turn forms another hydrogen bond with Tyr489 (RBD) (Fig. 2) (Table 4). Out of 16, in nine species (Figs. 3, 4, 5, 6) Gln24 is replaced by hydrophobic residue Leu24. Hence, there is a complete loss of the network of interactions (Table 4). Whereas, in M. musculus Gln is replaced by Asn and Tyr83 by Phe (Fig. 6). The interaction of Asn24 with Asn487 (RBD) was observed but the network of interactions was lost. In R. rattus, Gln24 is replaced by longer side chain residue Lys24 and Tyr83 replaced by Phe. Similarly, in R. rattus Lys24 form interaction with Asn487 but other interactions were not observed. In O. aries, Capra hircus, Bos taurus and B. bubalis Gln24 are conserved and the network of interactions was maintained (Fig. 4). The glycosylation site Asn90 and Asn103 also play important role in stabilizing the α1 helix interacting interface region 1 (Fig. 2). Interestingly, the Asn103 N-linked glycosylation is absent in all the species (Figs. 3, 4, 5, 6). In short, the Gln24 hydrogen-bonding network and the N-linked glycosylation sites Asn90 and Asn103 form a unique SARS-CoV-2 spike protein-interacting interface, which is absent in most of the species.
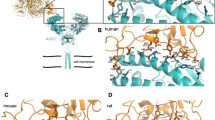
The ACE2 receptor-interacting surface of high to moderate susceptibility groups. a Felis catus (high); b Equus caballus (high); c Panthera tigris altaica (high); d Oryctolagus cuniculus (moderate). The hydrogen-bonded and hydrophobic interactions formed between region 1 to region 3 with spike protein are shown. The natural substitution of residues is colored pink and yellow. In region 1 Q24L mutation in all four species disrupts the Q24-N487 hydrogen bond. In region 2, despite D30E mutation in all four bond formation is intact. Similarly in region3 and region 4 all the hydrogen bonding is conserved. Glycosylation sites (Site1-5, Asn 53, Asn 90, Asn 103, Asn 322, Asn546, respectively) are marked in red and substitution of glycosylating residue colored in green
The ACE2 receptor-interacting surface of moderate to low susceptibility group. a Bos tauras; b Ovis aries; c Capra hircus; d Equus asinus. The hydrogen-bonded and hydrophobic interactions are shown from region 1 to region 3 and the natural substitution of residues are colored pink and yellow. In region 1 Q24L mutation disrupts the Gln24-Asn487 hydrogen bond in all four species and Tyr83Phe mutation disrupts the Tyr83-Asn487 bond in Equus asinus. In region 2 and region 3 hydrogen bonding is conserved despite residue substitution. Glycosylation sites (Site1-5, Asn 53, Asn 90, Asn 103, Asn 322, Asn546, respectively) are marked in red and substitution of glycosylating residue colored in green
The ACE2 receptor-interacting surface of low susceptibility group. The ACE2 receptor-interacting surface of a Bubalus bubalis; b Camelus dromedarius; c Canis lupus familiaris; d Ailuropoda melanoluca (very low). The hydrophobic and hydrogen-bonded interactions are shown from region 1 to region 3 and the natural substitution of residues are colored as pink and yellow. In region 1 Gln24Asn (Camelus dromedarius) and Gln24Leu (other three) substitution disrupts Gln24-Asn487 hydrogen bond. In region 2 His34Ser/Gln/Tyr mutation (in Bubalus bubalis, Camelus dromedarius and Ailuropoda melanoluca) disrupt the bond with Tyr453 while only in Canis lupus familiaris the bond is conserved. Lys31Asn mutation disrupts a major hydrogen bonding in Camelus dromedarius. Glycosylation sites (Site1-5, Asn 53, Asn 90, Asn 103, Asn 322, Asn546, respectively) are marked in red and substitution of glycosylating residue colored in green
The ACE2 receptor-interacting surface of very low susceptibility group. The ACE2 receptor-interacting surface of a Loxodonta africana; b Mus musculus; c Sus scrofa; d Rattus rattus along with hydrogen-bonded interactions formed with spike protein are shown from region 1 to region 3 along with the natural substitution of residues colored as pink and yellow. In region 1 Gln24-Asn487 hydrogen bond is disrupted due to Gln24Leu/Asn mutation. In region 2 His34-Gln493 is disrupted due to His34Tyr/Leu/Gln mutation in Mus musculus, Sus scrofa and Rattus rattus. Glycosylation sites (Site1-5, Asn 53, Asn 90, Asn 103, Asn 322, Asn546, respectively) are marked in red and substitution of glycosylating residue colored in green
SARS-CoV-2 spike protein-interacting region 2: mid portion of α1 helix of ACE2 receptor
The SARS-CoV-2 spike protein-interacting region 2 contains two hydrogen bond networks formed by Asp30/His34 and Lys31/Glu35. In the first network, the Asp30 OD1 atom forms a hydrogen bond with Lys417 (RBD) and Asp30 (OD2) forms a salt bridge with His34 to position the residue to interact with Tyr453 (RBD). In the second network, Lys31 and Glu35 both form hydrogen-bonded interactions with Gln493 (RBD) OE1 and NE2, respectively, along with an intra-network salt bridge (Fig. 2). These two networks of hydrogen-bonded interactions hold the central part of the α1 helix to form tight interaction with SARS-CoV-2 spike protein (Table 4, Fig. 2). In M. musculus, Asp30, Lys31 is replaced by Asn and His34 replaced by Gln. Hence, only the second network of interactions with Gln493 (RBD) is observed in the complex (Fig. 6). In Loxodonta Africana, Lys31 is replaced by Thr and His34 by Gln as a result no network of interactions observed but individual interaction of Asp30 with Lys417(RBD), Gln34 with Tyr453(RBD) and Glu35 with Gln493(RBD) could be observed (Fig. 6). In R. rattus, only the second network of interactions Gln493(RBD) could be observed (Fig. 6). In Ailuropoda melanoleuca, Canis lupus familiaris, O. cuniculus, E. caballus, E. asinus and S. scrofa, Asp30 replaced by Glu and His34 is replaced by Tyr/Gln/Ser/Leu. In these species, the first network of interactions could not be observed while the second network is maintained (Figs. 3, 4, 5, 6). Although individual residue interactions such as Glu30 with Lys417(RBD) were observed. In B. bubalis, Panthera tigris altaica, Felis catus, O. aries, Capra hircus, Bos taurus, the residue Asp30 is substituted with Glu but both the network of interactions could be observed (Figs. 4, 5, 6). In Camelus dromedarius, Lys31 is replaced by Glu hence the second network of interactions was not observed (Fig. 5). Consistently it is noted that both networks of interactions were not observed in all the species, either the Asp30 network or Lys31 network was observed. Although in many models individual interactions with the SARS-CoV-2 spike protein residues Lys417 and Tyr453 were observed. Hence, the two interchain and intrachain hydrogen bonding interactions and hydrophobic interactions required by SARS-CoV-2 spike protein to recognize the ACE2 receptor more efficiently.
SARS-CoV-2 spike protein-interacting region 3: end portion of α1 helix of ACE2 receptor
The SARS-CoV-2 spike protein-interacting region 3 is formed by the end portion of the α1 helix and the loop connecting β3 and β4 strands of the human ACE2 receptor. In this region, two hydrogen-bonding network was observed between the two proteins. The first network involves residues Asp38, Gln42 from α1helix and Lys353 from β3-β4. The Asp38 OD1 atom form a salt bridge with Lys353 and the Asp (OD2) forms a hydrogen bond with Tyr449 (RBD). Similarly, the Gln42 also forms a hydrogen bond with Tyr449 (RBD) and the backbone atom of Gly446 (RBD). The salt bridge between Asp38 and Lys353 holds the loop connecting β3 and β4 strands close enough that the backbone carbonyl oxygen atom of Lys353 forms a hydrogen bond with the backbone amide of Gly502 of the receptor-binding domain. The second hydrogen-bonded network involves residues Tyr41 from the α1 helix, Asp355 and Arg357 from the β4 strand. The Tyr41 forms hydrogen-bonded interaction with Thr500 (RBD) and Asp355. The Asp355 is stabilized by a salt bridge formed by Arg357 from the β4 strand (Fig. 2). The residue Asp38 is replaced by Glu in Canis lupus familiaris, E. asinus, E. caballus, Felis catus and Panthera tigris altaica. Hence, the first network of interaction with Lys353 was not observed in these species. But the interaction between Glu38 and Tyr449 (RBD) was observed. The second hydrogen-bonded network was maintained in these species. In all other species, no residue replacement was observed hence both the network of hydrogen-bonded was maintained in all models (Figs. 3, 4, 5, 6, Table 4).
In the vicinity of the region 3 interacting interface two glycosylation sites, Asn53 (site1) and Asn322 (site4) are present. The two hydrogen-bonding networks along with two glycosylations Asn322 and Asn53 help spike 2 protein to recognize the human receptor (Fig. 2). The residue Asn53 is conserved in all the species. In contrast, the N-linked glycosylation site Asn322 is absent in O. cuniculus, Bos taurus, Ovis aries, Capra hircus, Bubalus bubalis Loxodonta Africana, M. musculus and R. rattus (Table 3).
SARS-CoV-2 spike protein-interacting region 4: end portion of α2 helix of ACE2 receptor
The SARS-CoV-2 spike protein-interacting region 4 is formed by the end portion of the α2 helix. In this region, one hydrogen bond formed between Tyr83 and Tyr489 of spike RBD along with hydrophobic interaction with the aromatic residue Phe483 of SARS-CoV-2 spike sandwiched in between Met82 and Tyr83 of the receptor. The Met82 is crucial in terms of both hydrophobic interaction and holding the glycosylation site. In this region, the N-linked glycosylation Asn103 might play a crucial role in holding the helix α2 (Fig. 2). In the sugar modeled structure, we could observe that the residues Met82 and Gln81 hold the sugar moieties at the N-linked glycosylated site Asn103. In all species, Met82 is replaced with either Thr/Ser/Asn/Asp and the N-linked glycosylation site Asn103 is absent. In M. musculus, R. rattus and Loxodonta Africana, even the residue Tyr83 is substituted by Phe hence the hydrogen-bonded is also not observed. Hence, the entire network of hydrophobic interaction and N-linked glycosylation is absent in all species (Figs. 1, 3, 4, 5, 6).
Significant hydrophobic interactions
The SARS-CoV-2 spike protein-interacting regions in the human ACE2 receptor is majorly formed by α1 and α2 helices. Both the helices are held together majorly by the hydrophobic interactions formed between them. The hydrophobic core is formed by the residues Phe28, Phe32, Phe40, Trp69, Phe72, Leu79, Tyr83, Leu100, Phe390 and Leu391 (Table 5). These residues are evenly distributed from left to right of the α helices. Overall the hydrophobic residues give considerable stability to the arch-shaped spike interacting region in the ACE2 receptor. Apart from the hydrophobic core, there are few noteworthy hydrophobic interactions formed between SARS-CoV2 spike protein and the ACE2 receptor such as Thr27–Phe456, Tyr41–Gln498, Leu79–Phe486, Met82–Phe486, Tyr83–Phe486 and Lys353–Tyr505. The Left-hand portion of the concave interface is supported by Leu79–Phe486, Met82–Phe486, Tyr83–Phe486. The middle part and the right-hand side of the interface is held up by Tyr41–Gln498 and Lys353–Tyr505 sequentially (Table 4). Notably, in other mammalian species, many hydrophobic residues are naturally replaced by polar/charged residues such as Phe40 to Ser and Leu79 to Thr/His. All the residue substitutions are shown in (Table 4).
Discussion
The trimeric SARS-CoV-2 glycosylated spike protein binds to the glycosylated ACE2 receptor by interacting with its metalloprotease N-terminal domain (Lan et al. 2020). The arch-shaped spike protein binding interface in the human ACE2 receptor can be divided into four interacting regions. To form a stable complex the spike protein needs to maintain at least two hydrogen-bonded or salt bridge interactions at each site. Apart from these interactions, the hydrophobic core residues between α1 and α2 helices also play a crucial role in maintaining the ACE2 receptor to form an arch-shaped interacting surface for SARS-CoV-2 spike protein recognition (Brooke and Prischi 2020; Lan et al. 2020). Any substitution in these residues affects spike protein binding (Shi et al. 2020). Similarly, the Lance Wells group performed an MD simulation of glycosylated SARS-CoV-2 spike protein with the glycosylated human ACE2 receptor highlighting the importance of glycan-protein interactions to form a stable complex (Zhao et al. 2020a). In their structure model, they have shown that the sugar moieties in the N-linked glycosylation site Asn53, Asn90, Asn103, Asn322, and Asn546 of ACE2 receptor interacts with SARS-CoV-2 spike protein glycosylation sites N0074, N0165 and N0343. By including both studies (Ji et al. 2020; Shi et al. 2020; Zhao et al. 2020a), we analyzed the structure complex for the presence of conserved hydrogen bonds, hydrophobic interactions and the number of active glycosylation sites between the ACE2 receptor of 16 mammalian species with SARS-CoV-2 spike protein. The calculated Gibbs free energy (∆G) based on the interactions between ACE2 receptor and SARS-CoV-2 spike protein showed Felis catus having lowest free energy (− 98.04 kcal/mol) with four glycosylation site and the R. rattus (− 43.21 kcal/mol) with highest free energy having only two glycosylation sites (Tables 4, 6). The free binding energy indicates that there is a loss of some crucial interactions. Upon comparing the surface potential of human ACE2 receptor with other mammalian species, it clearly showed that substitution of residues Gln24, His34, Phe40 and Met82 at the N-terminal α1 helix and α2 helix results in a change in charge distribution at the interacting site (Fig. 7). Overall, five glycosylation sites and the crucial hydrogen-bonded and hydrophobic interactions make the human ACE2 receptor more specific than other mammalian species. Based on the analysis, we could categorize 16 species as high, moderate, low and very low susceptible species for SARS-CoV-2 infection. The Felis catus, E. caballus and Panthera tigris altaica are categorized as highly susceptible (Shi et al. 2020) as they lack two networks of interactions and one N-linked glycosylation site Asn103 (Table 3–6). The species O. cuniculus, B. Tauras, O. aries and C. hircus as moderately susceptible as they lack two networks of interactions and two N-linked glycosylation sites at Asn103 and Asn322. In the low susceptibility category, E. asinus, B. bubalis, Canis lupus familiaris, and Ailuropoda melaleuca as species lack three networks of interactions and one or two N-linked glycosylation sites Asn103 and Asn322. The species Loxodonta Africana, M. musculus, S. scrofa and R. rattus as least susceptible due to the absence of three networks of interactions and three N-linked glycosylation sites Asn90, Asn103 and Asn322. Although there is the loss of a crucial network of interactions, individual interactions were observed between SARS-CoV-2 spike protein and the ACE2 receptor. To form a stable complex the glycan moieties of SARS-CoV-2 spike protein interact with the glycans of the ACE2 receptor (Han et al. 2007b; Shajahan et al. 2020b; Zhao et al. 2020a). Hence, the glycan-glycan interaction between ACE2 receptor and SARS-CoV-2 spike protein could contribute to the susceptibility for infections. This is the first study to be reported, where 16 different mammalian species were categorized for susceptibility to infection based on the structural molecular interactions and the presence of active glycosylation sites in the ACE2 receptor. Our study correlates well with a previously reported study where cats and ferrets are reported as highly susceptible compared to dogs, pigs and mice (Rangel et al. 2020). However, the study reported by Shen et al. (Shen et al. 2020) described both cats and dogs are prone to infection than other species in their study. Our sequence and structure-based interaction analysis shows that cat, horse, rabbit and cattle are more prone to infection than dogs. Overall, our results contribute to explain the susceptibility of infection in various species based on residue substitution and the presence of N-linked glycosylation sites and found cats and horses being highly susceptible while wild bears and black rats as least susceptible to infection.
Conclusion
Our study shows that natural substitution at crucial interacting regions and the loss of N-linked glycosylation sites in the ACE2 receptors makes animals less susceptible to infection, with no primary role in transmission. Based on our analysis, F. catus, E. caballus, Panthera tigris altaica are highly susceptible while O. cuniculus, B. Tauras, O. aries and Capra hircus are moderately susceptible. The species E. asinus, B. bubalis, Canis lupus familiaris, Ailuropoda melaleuca and Camelus dromedarius are categorized as low susceptible with Loxodonta Africana, M. musculus, Sus scrofa and R. rattus as least susceptible species for infection. The difference in the susceptibility and host pathogenesis is also due to the loss of N-linked glycosylation sites and crucial interactions play an important role in the recognition and binding affinity between SARS-CoV-2 spike protein and ACE2 receptor. Our study also revealed that all the mammalian species except human lack the Asn103 N-linked glycosylation site and some species lack upto three glycosylation sites that play a key role in SARS-CoV2 spike protein recognizing the ACE2 receptor. However, more experimental and clinical studies and epidemiological reports are required to understand the host infection in the animals.
References
Almendros A, Gascoigne E (2020) Can companion animals become infected with Covid-19? Veterinary Record
Baloch S, Baloch MA, Zheng T, Pei X (2020) The coronavirus disease 2019 (COVID-19) pandemic. Tohoku J Experiment Med
Banerjee A, Kulcsar K, Misra V, et al (2019) Bats and coronaviruses. Viruses
Beard H, Cholleti A, Pearlman D et al (2013) Applying physics-based scoring to calculate free energies of binding for single amino acid mutations in protein-protein complexes. PLoS ONE 8:e82849. https://doi.org/10.1371/journal.pone.0082849
Bhowmik D, Pal S, Lahiri A et al (2020) Emergence of multiple variants of SARS-CoV-2 with signature structural changes. BioRxiv. https://doi.org/10.1101/2020.04.26.062471
Brooke GN, Prischi F (2020) Structural and functional modelling of SARS-CoV-2 entry in animal models. Sci Rep 10:15917. https://doi.org/10.1038/s41598-020-72528-z
Cevik M, Bamford CGG, Ho A (2020) COVID-19 pandemic—a focused review for clinicians. Clin Microbiol Infec
Chen Y, Liu Q, Guo D (2020) Emerging coronaviruses: Genome structure, replication, and pathogenesis. J Med Virol
Colovos C, Yeates TO (1993) Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci 2:1511–1519. https://doi.org/10.1002/pro.5560020916
Contini C, Di NM, Barp N et al (2020) The novel zoonotic COVID-19 pandemic: An expected global health concern. J Infec Develop Count. https://doi.org/10.3855/jidc.12671
Donoghue M, Hsieh F, Baronas E et al (2000) A novel angiotensin-converting enzyme-related carboxypeptidase (ACE2) converts angiotensin I to angiotensin 1–9. Circ Res. https://doi.org/10.1161/01.res.87.5.e1
Gabutti G, d’Anchera E, Sandri F, et al (2020) Coronavirus: Update Related to the Current Outbreak of COVID-19. Infec Dis Therapy
Han DP, Lohani M, Cho MW (2007) Specific asparagine-linked glycosylation sites are critical for DC-sign and L-sign-mediated severe acute respiratory syndrome coronavirus entry. J Virol. https://doi.org/10.1128/jvi.00315-07
Hasöksüz M, Kiliç S, Saraç F (2020) Coronaviruses and sars-cov-2. Turkish J Med Sci
Ji W, Wang W, Zhao X et al (2020) Cross-species transmission of the newly identified coronavirus 2019-nCoV. J Med Virol 92:433–440. https://doi.org/10.1002/jmv.25682
Jiang S, Hillyer C, Du L (2020) Neutralizing antibodies against SARS-CoV-2 and other human coronaviruses. Trends Immunol
Kannan S, Shaik Syed Ali P, Sheeza A, Hemalatha K (2020) COVID-19—recent trends. Europ Rev Med Pharmacol Sci https://doi.org/10.26355/eurrev_202002_20378
Lan J, Ge J, Yu J et al (2020) Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. https://doi.org/10.1038/s41586-020-2180-5
Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26:283–291. https://doi.org/10.1107/S0021889892009944
Levitt M, Gerstein M (1998) A unified statistical framework for sequence comparison and structure comparison. Proc Natl Acad Sci U S A 95:5913–5920. https://doi.org/10.1073/pnas.95.11.5913
Li W, Zhang C, Sui J et al (2005) Receptor and viral determinants of SARS-coronavirus adaptation to human ACE2. EMBO J 24:1634–1643. https://doi.org/10.1038/sj.emboj.7600640
Li R, Pei S, Chen B et al (2020) Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (SARS-CoV-2). Science. https://doi.org/10.1126/science.abb3221
Mousavizadeh L, Ghasemi S (2020) Genotype and phenotype of COVID-19: their roles in pathogenesis. J Microbiol Immunol Infect
Naz F, Mashkoor M, Sharma P, et al (2020) Drug Repurposing Approach to Target FtsZ Cell Division Protein From Salmonella Typhi. In: Intern J Biol Macromolecules https://pubmed.ncbi.nlm.nih.gov/32417543/. Accessed 24 Jun 2020
Ohtsubo K, Marth JD (2006) Glycosylation in cellular mechanisms of health and disease. Cell
Rangel HR, Ortega JT, Maksoud S et al (2020) SARS-CoV-2 host tropism: an in silico analysis of the main cellular factors. Virus Res 289:198154. https://doi.org/10.1016/j.virusres.2020.198154
Schwarz F, Aebi M (2011a) Mechanisms and principles of N-linked protein glycosylation. Curr Opin Struct Biol. https://doi.org/10.1016/j.sbi.2011.08.005
Schwarz F, Aebi M (2011b) Mechanisms and principles of N-linked protein glycosylation. Current Opinion Structural Biol
Shajahan A, Archer-Hartmann S, Supekar NT et al (2020) Comprehensive characterization of N and O-glycosylation of SARS-CoV-2 human receptor angiotensin converting enzyme 2. BioRxiv. https://doi.org/10.1101/2020.05.01.071688
Shang J, Wan Y, Luo C et al (2020) Cell entry mechanisms of SARS-CoV-2. Proc Natl Acad Sci USA. https://doi.org/10.1073/pnas.2003138117
Shen M, Liu C, Xu R, et al (2020) SARS-CoV-2 Infection of Cats and Dogs?
Shi J, Wen Z, Zhong G et al (2020) Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS-coronavirus 2. Science. https://doi.org/10.1126/science.abb7015
Shivakumar D, Williams J, Wu Y et al (2010) Prediction of absolute solvation free energies using molecular dynamics free energy perturbation and the OPLS force field. J Chem Theory Comput 6:1509–1519. https://doi.org/10.1021/ct900587b
Sievers F, Higgins DG (2014) Clustal Omega, accurate alignment of very large numbers of sequences. Methods Mol Biol 1079:105–116. https://doi.org/10.1007/978-1-62703-646-7_6
Singhal T (2020) A Review of Coronavirus Disease-2019 (COVID-19). Indian J Pediat
Stout AE, André NM, Jaimes JA et al (2020) Coronaviruses in cats and other companion animals: where does SARS-CoV-2/COVID-19 fit? Vet Microbiol 247:108777. https://doi.org/10.1016/j.vetmic.2020.108777
Temmam S, Barbarino A, Maso D et al (2020) Absence of SARS-CoV-2 infection in cats and dogs in close contact with a cluster of COVID-19 patients in a veterinary campus. One Health 10:100164. https://doi.org/10.1016/j.onehlt.2020.100164
Tipnis SR, Hooper NM, Hyde R et al (2000) A human homolog of angiotensin-converting enzyme: cloning and functional expression as a captopril-insensitive carboxypeptidase. J Biol Chem. https://doi.org/10.1074/jbc.M002615200
Towler P, Staker B, Prasad SG et al (2004) ACE2 X-ray structures reveal a large hinge-bending motion important for inhibitor binding and catalysis. J Biol Chem 279:17996–18007. https://doi.org/10.1074/jbc.M311191200
Waterhouse A, Bertoni M, Bienert S et al (2018) SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 46:W296–W303. https://doi.org/10.1093/nar/gky427
Yadav P, Kumar M, Bansal R et al (2019) Structure model of ferrochelatase from Salmonella typhi elucidating metalation mechanism. Int J Biol Macromol 127:585–593. https://doi.org/10.1016/j.ijbiomac.2019.01.066
Yan R, Zhang Y, Li Y et al (2020) Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. https://doi.org/10.1126/science.abb2762
Ye ZW, Yuan S, Yuen KS, et al (2020) Zoonotic origins of human coronaviruses. Intern J Biol Sci
Zhang T, Wu Q, Zhang Z (2020) Probable pangolin origin of 2019-nCoV associated with outbreak of covid-19. SSRN Electronic J. https://doi.org/10.2139/ssrn.3542586
Zhang T, Wu Q, Zhang Z (2020) Probable pangolin origin of SARS-CoV-2 associated with the covid-19 outbreak. Curr Biol. https://doi.org/10.1016/j.cub.2020.03.022
Zhao P, Praissman JL, Grant OC et al (2020) Virus-receptor interactions of glycosylated SARS-CoV-2 spike and human ACE2 receptor. Cell Host Microbe. https://doi.org/10.1016/j.chom.2020.08.004
Zhao WM, Song SH, Chen ML, et al (2020b) The 2019 novel coronavirus resource. Yi chuan = Hereditas https://doi.org/10.16288/j.yczz.20-030
Acknowledgments
The authors thank AIIMS Intramural funding for providing the software and hardware for the modeling studies.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by Tirthankar Koley, Shivani Madaan and Abdul S. Ethayathulla. The first draft of the manuscript was written by Tirthankar Koley, Shivani Madaan and Abdul S. Ethayathulla and all authors gave their inputs in the previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest with the current work or its publication.
Ethical approval
This article does not involve any study with human participants or animals to be performed by any authors.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Koley, T., Madaan, S., Chowdhury, S.R. et al. Structural analysis of COVID-19 spike protein in recognizing the ACE2 receptor of different mammalian species and its susceptibility to viral infection. 3 Biotech 11, 109 (2021). https://doi.org/10.1007/s13205-020-02599-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-020-02599-2