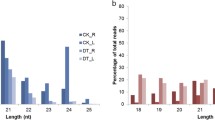
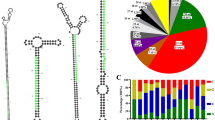
Abstract
MicroRNAs (miRNAs) play an important role in abiotic stress response in plants. However, the total miRNA profiles (miRNome) and drought-responsive miRNAs in H. tibetana have not been identified. In this study, we present the first report on the miRNome profiles of H. tibetana by high-throughput sequencing technology. 116 known and 4 predicted novel miRNAs were all identified in six H. tibetana samples. Moreover, to reveal the drought-responsive miRNAs in H. tibetana, we compared the miRNA profiles of H. tibetana grown under water sufficiency and drought stress. The results showed that 39 known miRNAs were up-regulated, while 34 miRNAs were downregulated under drought stress. Moreover, the expression of two novel miRNAs (novel_mir_24 and novel_mir_87) showed notable changes in response to drought stress. The target genes of these differentially expressed miRNAs were mainly enriched in cellular process, metabolic process, cell part, and response to stimulus. The identified drought-responsive miRNAs might be used for improving drought tolerance in H. tibetana and other plateau plants.




Similar content being viewed by others
References
Akdogan G, Tufekci ED, Uranbey S, Unver T (2016) miRNA-based drought regulation in wheat. Funct Integr Genomics 16:221–233
Atkinson NJ, Urwin PE (2012) The interaction of plant biotic and abiotic stresses: from genes to the field. J Exp Bot 63:3523–3543
Barik S, Sarkardas S, Singh A, Gautam V, Kumar P, Majee M, Sarkar AK (2014) Phylogenetic analysis reveals conservation and diversification of micro RNA166 genes among diverse plant species. Genomics 103:114–121
Barrera-Figueroa BE, Gao L, Diop NN, Wu Z, Ehlers JD, Roberts PA, Close TJ, Zhu JK, Liu R (2011) Identification and comparative analysis of drought-associated microRNAs in two cowpea genotypes. BMC Plant Biol 11:127
Candar-Cakir B, Arican E, Zhang B (2016) Small RNA and degradome deep sequencing reveals drought-and tissue-specific micrornas and their important roles in drought-sensitive and drought-tolerant tomato genotypes. Plant Biotechnol J 14:1727–1746
Chaudhary S, Sharma PC (2015) DeepSAGE based differential gene expression analysis under cold and freeze stress in seabuckthorn (Hippophae rhamnoides L.). PLoS ONE 10:e0121982
Cheah BH, Nadarajah K, Divate MD, Wickneswari R (2015) Identification of four functionally important microRNA families with contrasting differential expression profiles between drought-tolerant and susceptible rice leaf at vegetative stage. BMC Genomics 16:692
Chen M, Cao Z (2015) Genome-wide expression profiling of microRNAs in poplar upon infection with the foliar rust fungus Melampsora larici-populina. BMC Genomics 16:696
Fahlgren N, Howell MD, Kasschau KD, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Law TF, Grant SR, Dangl JL, Carrington JC (2007) High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of MIRNA genes. PLoS ONE 2:e219
Friedlander MR, Mackowiak SD, Li N, Chen W, Rajewsky N (2012) miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res 40:37–52
Ge A, Shangguan L, Zhang X, Dong Q, Han J, Liu H, Wang X, Fang J (2013) Deep sequencing discovery of novel and conserved microRNAs in strawberry (Fragaria x ananassa). Physiol Plant 148:387–396
Jones-Rhoades MW, Bartel DP, Bartel B (2006) MicroRNAS and their regulatory roles in plants. Annu Rev Plant Biol 57:19–53
Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, Yamanishi Y (2008) KEGG for linking genomes to life and the environment. Nucleic Acids Res 36:D480–D484
Kozomara A, Griffiths-Jones S (2014) miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 42:D68–D73
Li WX, Oono Y, Zhu J, He XJ, Wu JM, Iida K, Lu XY, Cui X, Jin H, Zhu JK (2008) The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance. Plant Cell 20:2238–2251
Morin RD, Aksay G, Dolgosheina E, Ebhardt HA, Magrini V, Mardis ER, Sahinalp SC, Unrau PJ (2008) Comparative analysis of the small RNA transcriptomes of Pinus contorta and Oryza sativa. Genome Res 18:571–584
Moxon S, Schwach F, Dalmay T, Maclean D, Studholme DJ, Moulton V (2008) A toolkit for analysing large-scale plant small RNA datasets. Bioinformatics 24:2252–2253
Mutum RD, Kumar S, Balyan S, Kansal S, Mathur S, Raghuvanshi S (2016) Identification of novel miRNAs from drought tolerant rice variety Nagina 22. Sci Rep 6:30786
Rogers K, Chen X (2013) Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell 25:2383–2399
Schwab R, Palatnik JF, Riester M, Schommer C, Schmid M, Weigel D (2005) Specific effects of microRNAs on the plant transcriptome. Dev Cell 8:517–527
Shinozaki K, Yamaguchi-Shinozaki K (2007) Gene networks involved in drought stress response and tolerance. J Exp Bot 58:221–227
Song C, Jia Q, Fang J, Li F, Wang C, Zhang Z (2010) Computational identification of citrus microRNAs and target analysis in citrus expressed sequence tags. Plant Biol (Stuttg) 12:927–934
Sunkar R, Zhou X, Zheng Y, Zhang W, Zhu JK (2008) Identification of novel and candidate miRNAs in rice by high throughput sequencing. BMC Plant Biol 8:1–17
Sunkar R, Li YF, Jagadeeswaran G (2012) Functions of microRNAs in plant stress responses. Trends Plant Sci 17:196–203
Wen M, Shen Y, Shi S, Tang T (2012) miREvo: an integrative microRNA evolutionary analysis platform for next-generation sequencing experiments. BMC Bioinform 13:140
Wu G, Park MY, Conway SR, Wang JW, Weigel D, Poethig RS (2009) The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138:750–759
Wu HJ, Ma YK, Chen T, Wang M, Wang XJ (2012) PsRobot: a web-based plant small RNA meta-analysis toolbox. Nucleic Acids Res 40:W22–W28
Xie Z, Khanna K, Ruan S (2010) Expression of microRNAs and its regulation in plants. Semin Cell Dev Biol 21:790–797
Xie C, Mao X, Huang J, Ding Y, Wu J, Dong S, Kong L, Gao G, Li CY, Wei L (2011) KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res 39:W316–W322
Zhang J, Xu Y, Huan Q, Chong K (2009) Deep sequencing of Brachypodium small RNAs at the global genome level identifies microRNAs involved in cold stress response. BMC Genomics 10:449
Zhang X, Gamarra J, Castro S, Carrasco E, Hernandez A, Mock T, Hadaegh AR, Read BA (2016) Characterization of the small RNA transcriptome of the marine Coccolithophorid, Emiliania huxleyi. PLoS One 11:e0154279
Zhou L, Chen J, Li Z, Li X, Hu X, Huang Y, Zhao X, Liang C, Wang Y, Sun L, Shi M, Xu X, Shen F, Chen M, Han Z, Peng Z, Zhai Q, Chen J, Zhang Z, Yang R, Ye J, Guan Z, Yang H, Gui Y, Wang J, Cai Z, Zhang X (2010) Integrated profiling of microRNAs and mRNAs: microRNAs located on Xq27.3 associate with clear cell renal cell carcinoma. PLoS ONE 5:e15224
Zhu QH, Helliwell CA (2011) Regulation of flowering time and floral patterning by miR172. J Exp Bot 62:487–495
Acknowledgements
The authors gratefully acknowledge the financial support from National Natural Science Foundation of China (No. 81473428) and the National Key Research and Development Program of China (No. 2017YFC1703900).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have declared there was no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Figure S1
Length distribution and abundance of the miRNome for the six Hippophae tibetana samples, i.e., HT-1, HT-2, HT-3, HT-4, HT-5, and HT-6 (PDF 427 kb)
Figure S2
Venn plot analysis of the predicted miRNAs for the six Hippophae tibetana samples, i.e., HT-1, HT-2, HT-3, HT-4, HT-5, and HT-6 (PDF 148 kb)
Table S1
Distribution of small RNAs among different categories in the six H. tibetana samples (HT-1, HT-2, HT-3, HT-4, HT-5, and HT-6) (XLSX 17 kb)
Table S2
Identified miRNAs as well as its expression profiles of the conserved miRNAs in the six H. tibetana samples (HT-1, HT-2, HT-3, HT-4, HT-5, and HT-6) (XLSX 58 kb)
Table S3
miRNAs expression profiles between HT123 (HT-1, HT-2 and HT-3) and HT456 (HT-4, HT-5 and HT-6) (XLSX 48 kb)
Table S4
KEGG pathway analysis of the target genes of differentially expressed miRNAs between HT123 (HT-1, HT-2 and HT-3) and HT456 (HT-4, HT-5 and HT-6) (XLSX 20 kb)
Table S5
Predicted novel miRNAs in the six H. tibetana samples (HT-1, HT-2, HT-3, HT-4, HT-5, and HT-6) (XLSX 22 kb)
Table S6
Expression difference of the novel miRNAs between HT123 (HT-1, HT-2 and HT-3) and HT456 (HT-4, HT-5 and HT-6) (XLSX 15 kb)
Table S7
Primers used for qRT-PCR (XLSX 15 kb)
Table S8
GO analysis (XLSX 10 kb)
Rights and permissions
About this article
Cite this article
Fan, G., Liu, Y., Du, H. et al. Identification of drought-responsive miRNAs in Hippophae tibetana using high-throughput sequencing. 3 Biotech 10, 53 (2020). https://doi.org/10.1007/s13205-019-2045-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-019-2045-5