Abstract
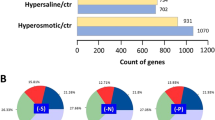
A high-salt-tolerant strain, Meyerozyma guilliermondii, has been isolated from activated sludge which has a great effect in the treatment of high-salt wastewater. To identify the salt tolerance mechanism of the strain, transcriptome sequencing and fluorescence quantitative PCR were used to analyse and compare the thallus, which were growing in the salt-free, low-salt (4%), and high-salt (16%) YPD media for 48 h, respectively, and then the change of intracellular and extracellular glycerol was determined. The results showed that 8220 unigenes were obtained from the sequencing data after de novo splicing, among which 6334 genes had annotation information in the SwissProt library. With salt-free as reference, there were 1135 unigenes that differentially expressed under low-salt stress and 1948 unigenes were differentially expressed under high-salt stress. With low salt as reference, 3056 unigenes were differentially expressed under high-salt stress. The fluorescence quantification results of the six candidate genes selected in this study were consistent with the transcriptome data. Under high-salt stress, the intracellular glycerol of M. guilliermondii reached a maximum value at about 10 min, then decreased quickly and tended to be stable. This study provided valuable data for the next study of salt tolerance of M. guilliermondii, and also contributed to the functional study of yeast salt tolerance genes.








Similar content being viewed by others
References
Albertyn J, Hohmann S, Thevelein JM et al (1994) GPD1, which encodes glycerol-3-phosphate dehydrogenase, is essential for growth under osmotic stress in Saccharomyces cerevisiae, and its expression is regulated by the high-osmolarity glycerol response pathway. Mol Cell Biol 14(6):4135–4144
Ansell Ricky, Granath Katarina, Hohmann Stefan et al (1997) The two isoenzymes for yeast NAD+-dependent glycerol 3-phosphate dehydrogenase encoded by GPD1 and GPD2 have distinct roles in osmoadaptation and redox regulation. EMBO J 16(9):2179
Blomberg A, Adler L (1989) Roles of glycerol and glycerol-3-phosphate dehydrogenase (NAD+) in acquired osmotolerance of Saccharomyces cerevisiae. J Bacteriol 171(2):1087–1092
Blomberg A, Adler L (1992) Physiology of osmotolerance in fungi. Adv Microb Physiol 33(33):145–212
Brown AD (1978) Compatible solutes and extreme water stress in eukaryotic micro-organisms. Adv Microb Physiol 17:181
Chen Raymond E, Thorner Jeremy (2007) Function and regulation in MAPK signaling pathways: lessons learned from the yeast Saccharomyces cerevisiae. BBA Mol Cell Res 1773(8):1311–1340
Cheng XZ, Wang ZX, Zhu GJ (2010) Progress in glycerol metabolism and its physiological function in yeast cells. China Biotechnol 30(5):140–148
Cronwright GR, Rohwer JM, Prior BA (2002) Metabolic control analysis of glycerol synthesis in Saccharomyces cerevisiae. Appl Environ Microbiol 68(9):4448
Davidson NM, Oshlack A (2014) Corset: enabling differential gene expression analysis for de novo assembled transcriptomes. Genome Biol 15(7):410
Fang J, Zeng KM (2005) Development of salt-containing wastewater treatment research. Ind Water Treat 25(2):1–4
Foury F, Roganti T, Lecrenier N et al (1998) The complete sequence of the mitochondrial genome of Saccharomyces cerevisiae. FEBS Lett 440(3):325–331
Ge HL, Chen L (2011) Effect of sewage irrigation on some physiological-biochemical characteristics of soybean (Glycine max) seedling and microbial functional groups in soybean rhizosphere soil. Henan Nongye Kexue (Journal of Henan Agricultural Sciences) 40(8):92–94
Gori K, Mortensen HD, Arneborg N et al (2010) Expression of the GPD1, and GPP2, orthologues and glycerol retention during growth of Debaryomyces hansenii, at high NaCl concentrations. Yeast 22(15):1213–1222
Grabherr MG, Haas BJ, Yassour M et al (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29(7):644
Griffin TJ, Gygi SP, Ideker T et al (2002) Complementary profiling of gene expression at the transcriptome and proteome levels in Saccharomyces cerevisiae. Mol Cell Proteomics 1(4):323–333
Guo SS, Zhang PY, Qu Y (2009) Review on biological treatment of high salinity wastewater and its feasibility. Sichuan Huanjing (Sichuan Environment) 28(3):85–88
Hanabusa K, Kondo K, Takemoto K (2011) Optimization studies on production of a salt-tolerant protease from Pseudomonas aeruginosa strain BC1 and its application on tannery saline wastewater treatment. Braz J Microbiol 42(4):1506–1515
Hohmann Stefan (2002) Osmotic stress signaling and osmoadaptation in yeasts. Microbiol Mol Biol Rev 66(2):300–372
Kanehisa M, Araki M, Goto S et al (2008) KEGG for linking genomes to life and the environment. Nucleic Acids Res 36((Database issue)):480–484
Kusvuran S (2014) Antioxidative enzyme activities in the leaves and callus tissues of salt-tolerant and salt-susceptible melon varieties under salinity. Afr J Biotech 11(3):635–641
Larsson C, Morales C, Gustafsson L et al (1990) Osmoregulation of the salt-tolerant yeast Debaryomyces hansenii grown in a chemostat at different salinities. J Bacteriol 172(4):1769–1774
Larsson C, Påhlman IL, Ansell R et al (1998) The importance of the glycerol 3-phosphate shuttle during aerobic growth of Saccharomyces cerevisiae. Yeast 14(4):347
Lei Y, Jie QL, Li YH (2007) The study on the treatment of high salinity wastewater. Environ Sci Manag 32(6):94–98
Li B, Dewey CN (2011) RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform 12(1):323
Maeda T, WurglerMurphy SM, Saito H (1994) A two-component system that regulates an osmosensing MAP kinase cascade in yeast. Nature 369(6477):242–245
Meikle AJ, Reed RH, Gadd GM (1991) The osmotic responses of Saccharomyces cerevisiae in K(+)-depleted medium. FEMS Microbiol Lett 62(1):89–93
Min HC, Yun HP (1999) Growth of Pichia guilliermondii, A9, an osmotolerant yeast, in waste brine generated from kimchi production. Biores Technol 70(3):231–236
Nevoigt E, Stahl U (2010) Osmoregulation and glycerol metabolism in the yeast Saccharomyces cerevisiae. FEMS Microbiol Rev 21(3):231–241
Nguyen HT, Dieterich A, Athenstaedt K et al (2004) Engineering of Saccharomyces cerevisiae for the production of l-glycerol 3-phosphate. Metab Eng 6(2):155–163
Pavlik P, Simon M, Schuster T et al (1993) The glycerol kinase (GUT1) gene of Saccharomyces cerevisiae: cloning and characterization. Curr Genet 24(1–2):21–25
Posas F, Takekawa M, Saito H (1998) Signal transduction by MAP kinase cascades in budding yeast. Curr Opin Microbiol 1(2):175–182
Proft M, Serrano R (1999) Repressors and upstream repressing sequences of the stress-regulated ENA1 gene in Saccharomyces cerevisiae: bZIP protein Sko1p confers HOG-dependent osmotic regulation. Mol Cell Biol 19(1):537–546
Raman M, Chen W, Cobb MH (2007) Differential regulation and properties of MAPKs. Oncogene 26(22):3100–3112
Shilo M (ed) (1979) Strategies of microbial life in extreme environments. Life science research report 13. Dahlem Konferenzen. Verlag Chemie, Weinheim, New York
Singh KK, Norton RS (1991) Metabolic changes induced during adaptation of Saccharomyces cerevisiae, to a water stress. Arch Microbiol 156(1):38–42
Thorsen M, Di Y, Tängemo C et al (2006) The MAPK Hog1p modulates Fps1p-dependent arsenite uptake and tolerance in yeast. Mol Biol Cell 17(10):4400
Trapnell C, Williams BA, Pertea G et al (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28(5):511–515
Tunbladjohansson I, Adler L (2010) Effects of sodium chloride concentration on phospholipid fatty acid composition of yeasts differing in osmotolerance. FEMS Microbiol Lett 43(3):275–278
Wang YQ, Tao T (1994) Compatible solutes in microbial osmotic pressure regulation. Microbiol China 34(5):355–359
Wang ZX, Zhu GJ (1999) Osmotic regulation and glycerol metabolism in yeast. China Biotechnol 19(5):34–39
Woolard CR, Irvine RL (1995) Treatment of hypersaline wastewater in the sequencing batch reactor. Water Res 29(4):1159–1168
Young MD, Wakefield MJ, Smyth GK et al (2010) Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol 11(2):R14
Zhang SW (2014) The influence of different concentrations of domestic sewage on growth of seedlings of Kandelia candel. Jiamusi Jiaoyu Xueyuan Xuebao (Journal of Jiamusi Education Institute) 3:439–441
Zhang YD, Very AA, Wang LM et al (2011) A K+ channel from salt-tolerant melon inhibited by Na+. New Phytol 189(3):856–868
Zhang X, Zhou Y, Zhang N (2017) Short-term and long-term effects of Zn (II) on the microbial activity and sludge property of partial nitrification process. Biores Technol 228:315
Zhu GJ, Wang ZX (1999) The HOG pathway and glycerol synthesis in yeast. Acta Microbiol Sin 39(1):91–93
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant no. 31760449).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Yang, Hl., Liao, Yy., Zhang, J. et al. Comparative transcriptome analysis of salt tolerance mechanism of Meyerozyma guilliermondii W2 under NaCl stress. 3 Biotech 9, 286 (2019). https://doi.org/10.1007/s13205-019-1817-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-019-1817-2