Abstract
Nanobiotechnology is considered as one of the important branches of nanotechnology, and research on synthesis of nanoscale materials, silver in particular, using plant and plant parts has been progressing rapidly. Herein, we used bark extract of Alstonia scholaris one of the most important medicinal plants to synthesize silver nanoparticles (AgNPs) which exhibited excellent antimicrobial properties against biofilm formed in drinking water PVC pipes. The biosynthesis of silver nanoparticles was done by treating 90 mL of 1 mM AgNO3 aqueous solution with 10 mL of 5 % bark extract. As-prepared silver nanoparticles were characterized using the biophysical techniques such as UV–Vis spectroscopy, Fourier transform infrared spectroscopy, X-ray diffraction, thermogravimetric analysis, transmission electron microscopy, and dynamic light scattering for the measurement of hydrodynamic diameter and zeta potential. The kinetics of the antimicrobial activity against PVC biofilm of prepared silver nanoparticles were done using comparative solution suspension time-killing assessments and which are evidenced in Epi-fluorescent microscopic observations.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Nanobiotechnology, a branch of nanotechnology has been playing a decisive role in the twenty first century particularly in the fields of medicine, pharmaceuticals, and electronics. So far, the entire nanoscience was built on a basic scientific challenge of controlling the connections between the atoms at nanoscale. There are numerous methods reported by various researchers to synthesize nanoscale materials and out of which physical and chemical methods (Kruis et al. 2000; Sauto et al. 2012) were having inherent limitations up to a certain extent which impose an important hurdle in avenues of application, biology in particular, due to the arisen toxicity by the synthetic protocol. The present decade has witnessed rapid shift in nanoparticles synthesis strategies from physico-chemical methods to biological methods which use the biosources such as bacteria (Juibari et al. 2011), fungi (Fesharakil et al. 2010), and plants (Prabha et al. 2014) for the synthesis of nanoparticles. Further, it is relatively easier to design and produce nanoparticles using biological sources, such as plants and microorganisms (Dickerson et al. 2008; Mandal et al. 2006; Mohanpuria et al. 2008; Chen et al. 2009; Johnson et al. 2012; Unuma et al. 2011, Bhattacharya and Gupta 2005).
Use of higher plant extracts to treat infectious and non‐infectious diseases is an age-old practice. However, the scientific analysis of different natural sources for their possible medicinal potency is comparatively a recent origin (Skinner 1995). The emergence and spread of antibiotic resistance microorganisms have triggered this type of plant investigations (Cowan 1999). Higher plants can serve both as potential antimicrobial crude drugs as well as a source of new anti-infective agents (Rios and Reico 2005). A. scholaris (L.) R.Br. (Apocynaceae) is an evergreen tropical tree native to the Indian sub‐continent and South-East Asia having grayish rough bark and milky sap rich in poisonous alkaloid. The bark also called dita bark is traditionally used by many ethnic groups of North‐East India and also other parts of the world as a source of remedy against fungal infections, malarial fever, toothache, rheumatism, snake bite, dysentery, bowl disorder etc., and its latex is used in treating coughs and fever (Kumar 2002; Khanikar 2007). Among the several genera of Alstonia, only scholaris species has been studied for antimicrobial potency (Khan et al. 2003).
Nanoscale materials have emerged as novel antimicrobial agents owing to their high surface area-to-volume ratio and the unique chemical and physical properties, which increase their contact with microbes and their ability to permeate cells (Lamsal et al. 2011). Further, nanotechnology has amplified the effectiveness of silver nanoparticles as antimicrobial agents (Christensen et al. 2011). Silver is also the only material whose plasmon resonance can be tuned to any wavelength in the visible spectrum. Fundamental studies carried out in the last three decades indicated that silver nanoparticles exhibit a rare combination of valuable properties, namely, unique optical properties associated with the surface plasmon resonance (SPR), well-developed surfaces, catalytic activity, high electrical double-layer capacitance, etc. Nanosilver has been used extensively as an antibacterial agent in the health industry, food storage, textile coatings, and a number of environmental applications (Jancy Mary and Inbathamizh 2012).
Reports on using A. scholaris bark extract for the synthesis of metallic nanoparticles, silver in particular, are scant and the studies on antimicrobial kinetics of phytogenic silver nanoparticles are not been reported. Hence, in the present study we have used the bark extract of A. scholaris to synthesize silver nanoparticles which exhibited significant antibacterial effects and the kinetics of these antimicrobial effects have been reported for the first time. Further, structural and morphological characteristics of nanoparticles were investigated by X-ray diffraction (XRD), transmission electron microscopy (TEM), FTIR, UV–visible spectra, and thermogravimetric analysis (TGA). Epi-fluorescent microscopic studies reveal the effectiveness of nanoparticle-bacterial species interfacial contact with regard to bacterial killing.
Materials and methods
Materials
Silver nitrate (99 % pure) was purchased from Sigma Aldrich, India. Nutrient broth, Nutrient agar plate, fluorescein isothiocyanate (FITC), and propidium iodide (PI) were supplied by Hi-Media, India.
Sample collection (PVC Biofilm)
The PVC biofilm samples were collected from four different regions located in and around Tirupati, (Chittoor District) Andhra Pradesh, India. The samples were collected from bore water PVC pipelines and taken in the sterile container. The collected samples were in amorphous form and were stored in an ice box.
Collection of plant material
Healthy plant of A. scholaris was collected from Mumbai, Maharashtra, India. The identity of the plant was confirmed by Agarkar Research Institute, Pune, India. A voucher specimen (No. AHMA-23537) has been deposited for future reference. From the selected plant, bark was collected by scrapping the trunk using neat and clean knife during the month of May 2014 and collected material was carefully washed and dried at 45 °C to constant weight. The dried bark of plant material was powdered, passed through a BSS no. 85-mesh sieve, and stored in air-tight container.
Preparation of aqueous extract (AE)
The powdered plant material was extracted with distilled water by cold maceration. Powdered plant materials (10 g) were mixed with 100 mL of distilled water then the solution heated up to 80 °C for 1 h with continuous stirring. After that the extract was filtered using Whatman No.1 filter paper and stored at 4 °C for further use.
Biosynthesis of silver nanoparticles
Silver nanoparticles were made according to the recipe described in the literature (Creighton et al. 1979; Suh et al. 1983). Briefly, a 90-mL aqueous solution of 1.0 × 10−3 M silver nitrate was mixed with a 10-mL of 5 % aqueous solution of A. Scholaris bark extract. Triple distilled water was used for solutions and both solutions were chilled to ice temperature before mixing. By mixing both the solutions, the A. Scholaris Ag solution was yellow in color and the solution was stirred repeatedly for an hour where the color of the solution become dark brown and whose UV–Vis absorption was recorded as 440 nm. The initial concentration of the A. Scholaris silver nanoparticles was measured using ICP-OES and was found to be 170 ppm.
Measurement of concentration of AgNPs using inductively coupled plasma optical emission spectrophotometer (ICP-OES)
The concentrations of the A. Scholaris bark extract-mediated AgNPs were measured using inductively coupled plasma optical emission spectrophotometer (ICP-OES) (Prodigy XP, Leeman labs, USA). The samples were prepared with 10 times dilution after centrifugation at 4000 rpm (18 g) for 15 min. Then 20 mL of aliquot was loaded to the racks of automatic sampler and estimated the concentration of AgNPs thrice.
Characterization of silver nanoparticles
UV–Visible spectrum for synthesized nanoparticles
The UV–Vis spectrum of this solution was recorded in spectra 50 ANALYTIKJENA Spectrophotometer, which was operated in the wavelength range of 400–800 nm. The localized surface plasmon resonance (LSPR) of the silver nanoparticles was recorded and which is the characteristic UV–Vis absorbance of silver nanoparticles.
FTIR analysis for synthesized nanoparticles
The FTIR spectrum was taken in the mid IR region of 400–4000 cm−1. The spectrum was recorded using attenuated total reflectance technique. The sample was directly placed in the KBr crystal and the spectrum was recorded in the transmittance mode.
Particle size and zeta potential analyzer for synthesized nanoparticles
The aqueous suspension of the synthesized nanoparticles was filtered through a 0.22 μm syringe driven filter unit, and the hydro dynamic diameter (HDD) of the distributed nanoparticles was measured by the principle of dynamic light scattering (DLS) using Nanopartica (HORIBA, SZ-100) compact scattering spectrometer.
X-ray diffraction analysis for synthesized nanoparticles
The crystalline structure of the nanoparticles was determined using the XRD technique. The XRD pattern was recorded using computer controlled XRD-system, JEOL, and Model: JPX-8030 with CuKα radiation (Ni filtered = 13,418 Å) at the range of 40 kV, 20 A. The ‘peak search’ and ‘search match’ program built in software (syn master 7935) was used for the identification of XRD peak.
Transmission electron microscopy (TEM)
The surface morphology and size of the nanoparticles were studied using transmission electron microscopy (JEOL (JEM-1010)) with an accelerating voltage of 80 kV after drying of a drop of aqueous AgNO3 on the carbon-coated copper TEM grids. Samples were dried and kept under vacuum in desiccators before loading them onto a specimen holder. The particle size distribution of nanoparticles was evaluated using ImageJ 1.45 s software.
Thermogravimetric analysis (TGA)
The thermal behavior of silver nanoparticles was studied by TGA. (TA Instruments Model SDT Q600). The instrument was run from 30 to 800 °C and the yield of the product also recorded.
Killing efficiency
The Ag nanoparticles were divided into seven groups and one group (tube 1) was seeded with 0.5 mL nutrient broth, which served as a sterility control. Six groups (tubes 2–7) were seeded with 0.5 mL fresh mixed bacterial suspension culture at a concentration of 105 colony forming units per mL (cfu/mL) and added to 1.5 mL of Ag nanoparticles. After 20 min, 5 mL of saline was added to all tubes. These tubes were vortexed and incubated at 37 °C for 12 h. The reduction of bacterial efficiency was monitored by total viable counts at various time intervals (3, 6, and 12 h). 100 µL of sample was drawn from each of the seven groups, spread on to a nutrient agar plate, and incubated at 37 °C for 24 h for total viable counts. The percentage reduction in bacterial count was calculated by the formula.
The disruption of bacterial cell membrane with respect to incubation time was visualized by epi-fluorescence microscopy. FITC and propidium iodide (PI) dual stains were used for identifying living and dead cells, respectively. PI penetrates only damaged cells and binds to the DNA emitting red color, whereas FITC remains exterior to undamaged cell walls which lead to green emission. About 0.5 µL of dual stain (FITC–PI; 1:1%) was added to bacterial sample and then incubated for 15 min. The excess stain was rinsed with sterile distilled water and examined under Epi-fluorescence microscope (E200 Coolpix; Nikon, Tokyo, Japan).
Results and discussion
Selection of PVC biofilm (scale) bacterial strain
Drinking water pipeline bacterial species have unusual biological activities depending upon the different metabolisms under temperature, pH, and pressure. The major role of this mixed bacterial species is to convert biofilm to scale which is a major problem encountered in drinking water pipelines. Figure 1 depicts the pH of medium at regular intervals of time in the presence of bacterial species as compared with control (urea broth) under identical conditions. The pH curves of mixed bacteria in nutrient medium were plotted (initial pH of the medium was 6.5). In the presence of bacteria, the pH was 6.5 and was gradually increased up to 12 at 4th day and finally at the end of the 6th day gradually decreased to 8.0. It clearly indicates that the bacterium is an alkaliphilic organism. In the present investigation, mixed bacterial species isolated from drinking water PVC pipes were used to generate biofilm formation in the range of 6500 ppm at 5th day of incubation.
UV–Visible spectral analysis
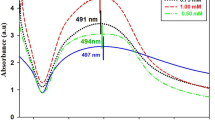
UV–Visible spectroscopy was employed to understand the biosynthesis of silver nanoparticles by A. scholaris. Fig. 2 shows the UV–Vis adsorption spectra of silver nanoparticles after 24 h of incubation at room temperature (37 °C). The spectrum shows peaks at 420–460 nm. But the maximum absorbance peak is observed at 440 nm which is the characteristic absorbance of AgNPs. The broadening of the absorption peak indicates the monodispersity of the formed silver nanoparticles.
FTIR analysis
FTIR spectrum is used to identify the possible chemical interactions among the silver salts and functional groups that are present in the extract. FTIR spectrum of the biosynthesized silver nanoparticles (Fig. 3) showed the absorption peaks at 1387.47, 1639.32, 2086.75, 3367.26 cm−1. The peak at 3367.26 and 2086.75 cm−1 reveals the presence of N–H bend, indicating the primary and secondary amine groups of protein. The band present at 568.19 shows the C–Br stretching likewise, the bands at 1639.32 and 1387.47 cm−1 correspond to the primary and secondary amine groups of N–H bending and carbonyl stretching vibrations of protein, respectively indicating the involvement of proteins in reduction and stabilization of silver ions and are bound to the functional organic groups (carboxyl and amine) that are present in the A. scholaris extract. These functional groups may act as template for reducing and capping of silver nanocrystals.
XRD analysis
The XRD patterns depicted in Fig. 4 demonstrate the crystalline nature of biosynthesized Ag nanoparticles. The sharp peaks with 2θ values of 37.9°, 44.1°, 64.3°, 77.3°, and 82.3° correspond to (111), (200), (220), (311), and (222) planes, respectively which confirms the face centered cubic (FCC) structure of the formed Ag nanoparticles. These Bragg’s reflections of silver were in good agreement with the JCPDS file No. 004-0783. The average crystalline size obtained from the Ag (111) diffraction line using Scherrer’s equation was 50 nm.
Dynamic light scattering analysis
DLS analysis measured HDD of the AgNO3 was found to be 55 nm (Fig. 5a). The recorded negative zeta potential of −10.4 mV with a single peak (Fig. 5b) signifies the presence of repulsive electro-static forces among the synthesized silver nanoparticles.
TGA analysis
TGA (Fig. 6) was conducted across the temperature range of 30–700 °C at a constant heating rate of 5 °C/min. The purity of the bioreduced silver nanoparticles was tested by TGA. It is evident from the obtained data that three distinct slopes of weight loss process at temperatures 99,200 and 445 °C were present. The curve shows the presence of OH groups at and below 200 °C and the curve at 445 °C was assigned to carbon contents which are present in the nanoparticles. The TGA shows that metal surface desorption of bio-organic substance (4–20 %) is present in the sample. The impurities could act as nucleation and capping agents for nanoparticles. TGA result shows that the purity of silver nanoparticles was 95 %.
TEM analysis
The size and shape of bioreductant nanoparticles were characterized and shown by the TEM micrograph of silver nanoparticles (Fig. 7). It is evident from the micrograph that individual silver nanoparticles as well as a number of aggregates are present and they are spherical in shape with an average particle size of 50 nm.
Antibacterial activity
AgNPs synthesized using A. scholaris could act as an inhibitor and serve to control the rate of bacterial proliferation. AgNPs loaded into bacterial cultures proved to be effective in terms of time-killing efficacy against a variety of pathogenic microorganisms (Dastjerdi and Montazer 2010; Fouda et al. 2013; Kiwi and Pulgarin 2010). In the present study, the time-killing efficacy of AgNPs loaded on nutrient agar medium was studied against mixed bacterial solution suspension. Ag nanoparticles were spread on the entire medium present in the petriplate, which generate reactive oxygen species (ROS) on the bacteria–nanoparticle interface, leading to bacterial cell wall destruction and resulted in decrease in the bacterial population. The killing of bacterial species in solution was monitored by bacterial solution suspension studies. Interfacial contact investigations were conducted in order to gain an insight on the interactions occurring at the Ag NPs–bacteria interface, and had provided substantial results with regard to bacterial killing as a result of consistent interfacial contact binding. Bacterial killing efficiency for mixed bacterial culture by solution suspension method is presented in Fig. 8. It was also observed that the mixed bacterial suspension survival after 7 h was very low(5 %) compared to 1 h (90 %), 2 h (80 %), 3 h (65 %), 4 h (45 %), 5 h (30 %), and 6 h (20 %).
The increase in time-killing efficacy with treatment may be due to the presence of AgNPs on bacterial surface at extended periods of time which is evidenced in the Epi-fluorescent microscopic images of mixed bacterial species under wet conditions. AgNPs samples were placed in the 4.5 × 105 CFU/mL of mixed bacterial culture at different time durations to render monitoring of both dead and living cells. Living cells were stained green in color, while the dead cells were stained with red color. The Epi-fluorescent microscopic images of mixed bacterial species were recorded at definite time intervals during bacterial killing by direct interfacial contact method. Hence, data on interfacial contact studies were recorded at 2-h intervals for mixed bacterial cultures Fig. 9a which depicts live bacterial cells stained in green. Initiation of breakdown of cell membrane is indicated by reddish regions around the bacterial species, as illustrated in Fig. 9b. The Epi-fluorescent image of bacterial cells after 4 h of AgNPs exposure leads to further deterioration of bacterial cells which may be due to the increased contact time as depicted in Fig. 9c and the same was observed even after 6 h. However, most cells of mixed bacterial species are completely destroyed after 7 h, as elucidated in Fig. 9d. It was found that complete nullification of bacterial cells occurred after time periods of 6 and 7 h, respectively, and this observation may be due to the fact that effective destruction of bacterial cells by reactive oxygen radical species and whose effectiveness could be a function of contact distance between AgNPs and bacterial cells. Hence, subdued bacterial killing rate in bacterial solution can be attributed to varying distances and inconsistent contact between the AgNPs and mixed bacterial species in solution. The destruction of bacterial species initiates at the cell membrane and rapidly proceeds along the interior of the cell, as depicted in the Epi-fluorescent microscopic images. AgNPs cause toxicity to the bacterial cell constituents, damage to mitochondria in particular, and result in depletion of ATP which enhances the production of ROS. Then there was a double impact of both ROS and AgNPs on the nucleus of bacterial cell leads to oxidation of DNA causing the death of the bacterial cell (Venkata Subbaiah et al. 2014). Similar results were observed by previous investigators for ZnO nanoparticles (Raghupathi et al. 2011; Sivakumar et al. 2010).
Conclusion
This contribution reported for the first time the killing efficacy of mixed bacterial cultures loaded with Ag nanoparticles synthesized using the extract of A. scholaris. XRD pattern has confirmed the FCC structure of formed Ag nanoparticles. TEM micrographs revealed the presence of spherical-shaped Ag nanoparticles with an average size of 50 nm. TGA studies show that purity of nanoparticles was 86 %. Further, the tensile force and maximum elongation at breakdown were greater for the nanoparticles loaded in bacterial medium indicating its mechanical strength and suitability for biofilm degradation and showing the anti-scaling property. AgNPs loaded on mixed bacterial suspension interfacial contact studies reveal substantial killing efficiency which may be due to the consistent interfacial contact between the bacterial species and nanoparticles. Thus, our studies once again confirmed the potential antimicrobial activity of phytogenic silver nanoparticles.
References
Bhattacharya D, Gupta RK (2005) Nanotechnology and potential of microorganisms. Crit Rev Biotechnol 25:199
Chen L, Shen Y, Xie A, Huang B, Jia R, Guo R (2009) Mediated synthesis of metal carbonate minerals with unusual morphologies and structures. Cryst Growth Des 9:743
Christensen L, Vivekanandhan S, Misra M, Mohanty AK (2011) Biosynthesis of silver nanoparticles using murraya koenigii (curry leaf): an investigation on the effect of broth concentration in reduction mechanism and particle size. Adv Mater Lett 2:429–434
Cowan MM (1999) Plant products as antimicrobial agents. Clin Microbiol Rev 12:564–582
Creighton JA, Blatchford CG, Albrecht MG (1979) Plasma resonance enhancement of Raman scattering by pyridine adsorbed on silver or gold sol particles of size comparable to the excitation wavelength. J Chem Soc Faraday Trans II 75:790–798
Dastjerdi R, Montazer M (2010) A review on the application of inorganic nanostructured materials in the modification of textiles: focus on anti-microbial properties. Colloids Surf B 79:5
Dickerson MB, Sandhage KH, Naik RR (2008) Protein-and peptide-directed syntheses of inorganic materials. Chem Rev 108:4935
Fesharakil PJ, Nazaril P, Shakibaiel M, Rezaiell S, Banoee M (2010) Biosynthesis of selenium nanoparticles using klebsiella pneumonia and their recovery by a simple sterilization process. Braz J Microbiol 41:461–466
Fouda MMG, Abdel-Halima ES, Al-Deyab SS (2013) Antibacterial modification of cotton using nanotechnology. Carbohydr Polym 92:943
Jancy Mary E, Inbathamizh L (2012) Green synthesis and characterization of nano silver using leaf extract of morinda pubescens. Asian J Pharm Clin Res 5:1
Johnson JM, Kinsinger N, Sun C, Li D, Kisailus D (2012) Urease-mediated room-temperature synthesis of nanocrystalline titanium dioxide. J Am Chem Soc 134:13974
Juibari MM, Abbaaalizadeh S, Jouzani GS, Noruzi M (2011) Intensified biosynthesis of silver nanoparticles using a native extremophilic ureibacillus thermosphaericus strain. Mater Lett 65:1014–1017
Khan MR, Omoloso AD, Kihara M (2003) Fitoterapia 74:736–740
Khanikar G (2007) Gharooa Sikitsher Nidan, 3rd edn. Puthiteertha Publication, Assam
Kiwi J, Pulgarin C (2010) Innovative bactericidal and self cleaning textiles. Catal Today 151:2
Kruis FE, Fissan H, Pellinghaus B (2000) Sintering and evaporation characteristics of gas-phase synthesis of size-selected pbs nanoparticles. Mater Sci Eng B 69:329–334
Kumar S (2002) Medicinal plants of North-East region, 1st edn. Scientific Publishers, Rajasthan
Lamsal K, Kim SW, Jung JH, Kim YS, Kim KS, Lee YS (2011) Inhibition effects of silver nanoparticles against powdery mildews on cucumber and pumpkin. Mycobiology 39:26–32
Mandal D, Bolander ME, Mukhopadhyay D, Sarkar G, Mukherjee P (2006) The use of microorganisms for synthesis of metal nanoparticles and their application. Appl Microbiol Biotechnol 69:485
Mohanpuria P, Rana NK, Yadav SK (2008) Biosynthesis of nanoparticles: technological concepts and future applications. J Nanopart Res 10:507
Prabha S, Supraja N, Garud M, Prasad TNVKV (2014) Synthesis, characterization and antimicrobial activity of Alstonia scholaris bark-extract-mediated silver nanoparticles. J Nanostruct Chem. doi:10.1007/s40097-014-0132-z
Raghupathi KR, Koodali RT, Manna AC (2011) Size-dependent bacterial growth inhibition and mechanism of antibacterial activity of zinc oxide nanoparticles. Langmuir 27:4020
Rios JL, Reico MC (2005) Medicinal plants and antimicrobial activity. J Ethnopharmacol 100:80–84
Sauto EB, Severino P, Santana MHA (2012) Preparation of polymeric nanoparticles by polymerization of monomers part-I. Pollmeros 22:96–100
Sivakumar PM, Balaji S, Prabhawathi V, Neelakandan R, Manoharan PT, Doble M (2010) Effective antibacterial adhesive coating on cotton fabric using ZnO nanorods and chalcone. Carbohydr Polym 79:717
Skinner FA (1995) Antibiotics. In: Paech K, Tracey MV (eds) Modern methods of plant analysis. Springer, Berlin, pp 626–654
Suh JS, DiLella DP, Muscovite M (1983) Surface-enhanced Raman spectroscopy of colloidal metal systems: two-dimensional phase equilibrium in p-aminobenzoic acid adsorbed on silver. J Phys Chem 87:1540–1544
Unuma H, Matsushima Y, Kawai T (2011) Enzyme-mediated synthesis of ceramic materials. J Ceram Soc Jpn 119:623
Venkata Subbaiah K, Susmila Aparna G, Sucharitha K, Sai Gopal DVR (2014) New generation of bactericidal silver nanoparticles against different antibiotic resistant Escherichia coli strains. Appl Nanosci. doi:10.1007/s13204-014-0381-7
Acknowledgments
The authors are thankful to the authorities of Acharya N G Ranga Agricultural University for providing research facilities at Institute of Frontier Technology, Regional Agricultural Research Station to carry out this research work.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Supraja, N., Prasad, T.N.V.K.V., David, E. et al. Antimicrobial kinetics of Alstonia scholaris bark extract-mediated AgNPs. Appl Nanosci 6, 779–787 (2016). https://doi.org/10.1007/s13204-015-0483-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13204-015-0483-x