Abstract
Background
Invasiveness is a very challenging clinical problem in nonfunctional pituitary adenomas (NFPAs), and currently, there are no effective invasiveness-related molecular biomarkers. The post-neurosurgery treatment is much different as for invasive and noninvasive NFPAs. The aim of this study was to integrate phosphoproteomics and transcriptomics data to reveal phosphorylation-mediated molecular events for invasive characteristics of NFPAs to achieve a potential tool for patient stratification, and prognostic/predictive assessment to discriminate invasive from noninvasive NFPAs for personalized attitude.
Methods
The 6-plex tandem mass tag (TMT) labeling reagents coupled with TiO2 enrichment of phosphopeptides and liquid chromatography-tandem mass spectrometry (LC-MS/MS) were used to identify and quantify each phosphoprotein and phosphosite in NFPAs and controls. Differentially expressed genes (DEGs) between invasive NFPA and control tissues were obtained from the Gene Expression Omnibus (GEO) database. The overlapping analysis was performed between phosphoprotiens and invasive DEGs. Gene Ontology (GO) enrichment, the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway, and protein–protein interaction (PPI) analyses were used to analyze these overlapped molecules.
Results
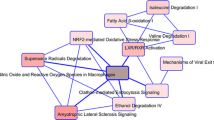
In total, 1035 phosphoproteins with 2982 phosphorylation sites were identified in NFPAs vs. controls, and 2751 DEGs were identified in invasive NFPAs vs. controls. Overlapping analysis of these phosphoproteins and DEGs exposed 130 overlapped molecules (phosphoproteins; invasive DEGs). GO enrichment and KEGG pathway analyses of 130 overlapped molecules revealed multiple biological processes and signaling pathway network alterations, including cell–cell adhesion, platelet activation, GTPase signaling pathway, protein kinase signaling, calcium signaling pathway, estrogen signaling pathway, glucagon signaling pathway, cGMP–PKG signaling pathway, GnRH signaling pathway, inflammatory mediator regulation of TRP channels, vascular smooth muscle contraction, and Fc gamma R-mediated phagocytosis, which were obviously associated with tumor invasive characteristics. For 130 overlapped molecules, PPI network-based molecular complex detection (MCODE) identified 10 hub molecules, namely SLC2A4, TSC2, AKT1, SCG3, ALB, APOL1, ACACA, SPARCL1, CHGB, and IGFBP5. These hub molecules are involved in multiple signaling pathways and represent potential predictive/prognostic markers in NFPA patients as well as they represent potential therapeutic targets.
Conclusions
This study provided the first large-scale phosphoprotein profiling and phosphorylation-related signaling pathway network alterations in human NFPA tissues. Further, overlapping analysis of phosphoproteins and invasive DEGs revealed the phosphorylation-mediated signaling pathway network changes in invasive NFPAs. These findings are the precious resource for in-depth insight into the molecular mechanisms of NFPAs, as well as for the discovery of effective phosphoprotein biomarkers and therapeutic targets for invasive NFPAs.



Similar content being viewed by others
Abbreviations
- ACACA:
-
acetyl-CoA carboxylase 1
- ACN:
-
acetonitrile
- AGC:
-
automatic gain control
- AKT:
-
protein kinase B
- AKT1:
-
RAC-alpha serine/threonine-protein kinase
- ALB:
-
serum albumin
- AMPK:
-
AMP-activated protein kinase
- APOL1:
-
apolipoprotein L1
- ATF2:
-
activating transcription factor 2
- BP:
-
biological process
- CC:
-
cellular component
- cGMP:
-
cyclic nucleotide cGMP
- CHGB:
-
secretogranin-1
- DEG:
-
differentially expressed gene
- DEP:
-
differentially expressed protein
- DTT:
-
dithiothreitol
- r-ERG channel:
-
rat ether-à-go-go-related (ERG) channel
- ERK:
-
extracellular signal–regulated kinase
- ESCRT:
-
endosomal sorting complex required for transport
- ESI-TRAP:
-
electrospray ionization-ion trap
- FC:
-
fold change
- FDR:
-
false discovery rate
- FGF-2:
-
fibroblast growth factor-2
- FPA:
-
functional pituitary adenoma
- GEO:
-
Gene Expression Omnibus
- GH:
-
growth hormone
- GH3:
-
pituitary growth hormone 3
- GnRH:
-
gonadotropin-releasing hormone
- GO:
-
Gene Ontology
- HCD:
-
high-energy collision dissociation
- HIF-1a:
-
hypoxia-inducible factor-1a
- HPLC:
-
high-performance liquid chromatography
- IGF-1:
-
insulin growth factor-1
- IGFBP5:
-
insulin-like growth factor-binding protein 5
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- LC:
-
liquid chromatography
- MAPK:
-
mitogen-activated protein kinase
- MCODE:
-
molecular complex detection
- MF:
-
molecular function
- MMPs:
-
matrix metalloproteinases
- MS/MS:
-
tandem mass spectrometry
- mTOR:
-
mammalian target of rapamycin
- NCBI:
-
National Center for Biotechnology Information
- NFκB:
-
nuclear factor kappa-B
- NFPA:
-
nonfunctional pituitary adenoma
- NMR:
-
nuclear magnetic resonance
- PACAP:
-
pituitary adenylyl cyclase activating polypeptide
- PI3K:
-
phosphatidylinositol 3 kinase
- PKG:
-
protein kinase G
- PPI:
-
protein–protein interaction
- PTM:
-
post-translational modification
- PTTG:
-
pituitary tumor-transforming gene
- RSK:
-
ribosomal S6 kinase
- SCG3:
-
secretogranin-3
- Ser or S:
-
serine
- SLC2A4:
-
solute carrier family 2 facilitated glucose transporter member 4
- S/N:
-
signal-to-noise
- SNAP:
-
soluble N-ethylmaleimide-sensitive fusion attachment protein
- SNARE:
-
soluble N-ethylmaleimide-sensitive factor attachment protein receptor
- SPARCL1:
-
SPARC-like protein 1
- TFA:
-
trifluoroacetic acid
- Thr or T:
-
threonine
- TMT:
-
tandem mass tag
- TSC2:
-
Tuberin
- TRH:
-
thyrotropin-releasing hormone
- Tyr or Y:
-
tyrosine
- VEGF:
-
vascular endothelial growth factor
References
Melmed S. Mechanisms for pituitary tumorigenesis: the plastic pituitary. J Clin Invest. 2003;112:1603–18. https://doi.org/10.1172/JCI20401.
Melmed S. Pathogenesis of pituitary tumors. Nat Rev Endocrinol. 2011;7:257–66. https://doi.org/10.1038/nrendo.2011.40.
Melmed S. Pituitary tumors. Endocrinol Metab Clin N Am. 2015;44:1–9. https://doi.org/10.1016/j.ecl.2014.11.004.
Zhan X, Desiderio DM. Editorial: Molecular network study of pituitary adenomas. Front Endocrinol. 2020;11:26. https://doi.org/10.3389/fendo.2020.00026.
Cheng T, Wang Y, Lu M, Zhan X, Zhou T, Li B, et al. Quantitative analysis of proteome in non-functional pituitary adenomas: clinical relevance and potential benefits for the patient. Front Endocrinol. 2019;10:854. https://doi.org/10.3389/fendo.2019.00854.
Wang Y, Cheng T, Lu M, Mu Y, Li B, Li X, et al. TMT-based quantitative proteomics revealed follicle-stimulating hormone (FSH)-related molecular characterizations for potentially prognostic assessment and personalized treatment of FSH-positive non-functional pituitary adenomas. EPMA J. 2019;10:395–414. https://doi.org/10.1007/s13167-019-00187-w.
Zhan X, Desiderio DM, Wang X, Zhan X, Guo T, Li M, et al. Identification of the proteomic variations of invasive relative to noninvasive nonfunctional pituitary adenomas. Electrophoresis. 2014;35(15):2184–94.
Losa M, Mortini P, Barzaghi R, Ribotto P, Terreni MR, Marzoli SB, et al. Early results of surgery in patients with nonfunctioning pituitary adenoma and analysis of the risk of tumor recurrence. J Neurosurg. 2008;108(3):525–32. https://doi.org/10.3171/JNS/2008/108/3/0525.
Meij BP, Lopes MB, Ellegala DB, Alden TD, Laws ER Jr. The long-term significance of microscopic dural invasion in 354 patients with pituitary adenomas treated with transsphenoidal surgery. J Neurosurg. 2002;96(2):195–208. https://doi.org/10.3171/jns.2002.96.2.0195.
Selman WR, Laws ER Jr, Scheithauer BW, Carpenter SM. The occurrence of dural invasion in pituitary adenomas. J Neurosurg. 1986;64(3):402–7. https://doi.org/10.3171/jns.1986.64.3.0402.
Cheng T, Zhan X. Pattern recognition for predictive, preventive, and personalized medicine in cancer. EPMA J. 2017;8:51–60. https://doi.org/10.1007/s13167-017-0083-9.
Grech G, Zhan X, Yoo BC, Bubnov R, Hagan S, Danesi R, et al. EPMA position paper in cancer: current overview and future perspectives. EPMA J. 2015;6:9. https://doi.org/10.1186/s13167-015-0030-6.
Zhan X, Desiderio DM. The use of variations in proteomes to predict, prevent, personalize treatment for clinically non-functional pituitary adenomas. EPMA J. 2010;1:439–59. https://doi.org/10.1007/s13167-010-0028-z.
Hu R, Wang X, Zhan X. Multi-parameter systematic strategy for predictive, preventive, and personalized medicine in cancer. EPMA J. 2013;4:2. https://doi.org/10.1186/1878-5085-4-2.
Lu M, Zhan X. The crucial role of multiomic approach in cancer research and clinically relevant outcomes. EPMA J. 2018;9(1):77–102. https://doi.org/10.1007/s13167-018-0128-8.
Zhan X, Long Y, Lu M. Exploration of variations in proteome and metabolome for predictive diagnostics and personalised treatment algorithms: innovative approach and examples for potential clinical application. J Proteome. 2018;188:30–40. https://doi.org/10.1016/j.jprot.2017.08.020.
Zhan X, Li B, Zhan X, Schlüter H, Jungblut PR, Coorssen JR. Innovating the concept and practice of two-dimensional gel electrophoresis in the analysis of proteomes at the proteoform level. Proteomes. 2019;7(4):36. https://doi.org/10.3390/proteomes704003.
Guo T, Wang X, Li M, Yang H, Li L, Peng F, et al. Identification of glioblastoma phosphotyrosine-containing proteins with two-dimensional Western blotting and tandem mass spectrometry. Biomed Res Int. 2015;2015:134050.
Singh V, Ram M, Kumar R, Prasad R, Roy BK, Singh KK. Phosphorylation: implications in cancer. Protein J. 2017;36:1–6. https://doi.org/10.1007/s10930-017-9696-z.
Golden RJ, Chen B, Li T, Braun J, Manjunath H, Chen X, et al. An argonaute phosphorylation cycle promotes microRNA-mediated silencing. Nature. 2017;542:197–202. https://doi.org/10.1038/nature21025.
Tsai CF, Wang YT, Yen HY, Tsou CC, Ku WC, Lin PY, et al. Large-scale determination of absolute phosphorylation stoichiometries in human cells by motif-targeting quantitative proteomics. Nat Commun. 2015;6:6622. https://doi.org/10.1038/ncomms7622.
Sergina NV, Rausch M, Wang D, Blair J, Hann B, Shokat KM, et al. Escape from HER-family tyrosine kinase inhibitor therapy by the kinase-inactive HER3. Nature. 2007;445(7126):437–41.
Shah KN, Bhatt R, Rotow J, Rohrberg J, Olivas V, Wang VE, et al. Aurora kinase A drives the evolution of resistance to third-generation EGFR inhibitors in lung cancer. Nat Med. 2019;25(1):111–8. https://doi.org/10.1038/s41591-018-0264-7.
Kreuzer J, Edwards A, Haas W. Multiplexed quantitative phosphoproteomics of cell line and tissue samples. Methods Enzymol. 2019;626:41–65. https://doi.org/10.1016/bs.mie.2019.07.027.
Li Z, Li M, Li X, Xin J, Wang Y, Shen QW, et al. Quantitative phosphoproteomic analysis among muscles of different color stability using tandem mass tag labeling. Food Chem. 2018;249:8–15. https://doi.org/10.1016/j.foodchem.2017.12.047.
Carretero L, Llavona P, López-Hernández A, Casado P, Cutillas PR, de la Peña P, et al. ERK and RSK are necessary for TRH-induced inhibition of r-ERG potassium currents in rat pituitary GH3 cells. Cell Signal. 2015;27(9):1720–30. https://doi.org/10.1016/j.cellsig.2015.05.014.
Zhao S, Feng J, Li C, Gao H, Lv P, Li J, et al. Phosphoproteome profiling revealed abnormally phosphorylated AMPK and ATF2 involved in glucose metabolism and tumorigenesis of GH-PAs. J Endocrinol Investig. 2019;42(2):137–48. https://doi.org/10.1007/s40618-018-0890-4.
Delcourt N, Thouvenot E, Chanrion B, Galéotti N, Jouin P, Bockaert J, et al. PACAP type I receptor transactivation is essential for IGF-1 receptor signalling and antiapoptotic activity in neurons. EMBO J. 2007;26(6):1542–51.
Beranova-Giorgianni S, Zhao Y, Desiderio DM, Giorgianni F. Phosphoproteomic analysis of the human pituitary. Pituitary. 2006;9(2):109–20.
Long Y, Lu M, Cheng T, Zhan X, Zhan X. Multiomics-based signaling pathway network alterations in human non-functional pituitary adenomas. Front Endocrinol. 2019;10:835. https://doi.org/10.3389/fendo.2019.00835.
Ota M, Gonja H, Koike R, Fukuchi S. Multiple-localization and hub proteins. PLoS One. 2016;11:e0156455. https://doi.org/10.1371/journal.pone.0156455.
Zhan X, Li N, Zhan X, Qian S. Revival of 2DE-LC/MS in proteomics and its potential for large-scale study of human proteoforms. Med One. 2018;3:e180008. https://doi.org/10.20900/mo.20180008.
Zhan X, Yang H, Peng F, Li J, Mu Y, Long Y, et al. How many proteins can be identified in a 2-DE gel spot within an analysis of a complex human cancer tissue proteome? Electrophoresis. 2018;39:965–80. https://doi.org/10.1002/elps.201700330.
Aebersold R, Agar JN, Amster IJ, Baker MS, Bertozzi CR, Boja ES, et al. How many human proteoforms are there? Nat Chem Biol. 2018;14(3):206–14. https://doi.org/10.1038/nchembio.2576.
Smith LM, Kelleher NL. Proteoforms as the next proteomics currency. Science. 2018;359(6380):1106–7. https://doi.org/10.1126/science.aat1884.
Broncel M, Treeck M. Label-based mass spectrometry approaches for robust quantification of the phosphoproteome and total proteome in Toxoplasma gondii. Methods Mol Biol. 2020;2071:453–68. https://doi.org/10.1007/978-1-4939-9857-9_23.
Serioli S, Doglietto F, Fiorindi A, Biroli A, Mattavelli D, Buffoli B, et al. Pituitary adenomas and invasiveness from anatomo-surgical, radiological, and histological perspectives: a systematic literature review. Cancers (Basel). 2019;11(12). https://doi.org/10.3390/cancers11121936.
Zheng X, Li S, Zhang W, Zang Z, Hu J, Yang H. Current biomarkers of invasive sporadic pituitary adenomas. Ann Endocrinol (Paris). 2016;77(6):658–67. https://doi.org/10.1016/j.ando.2016.02.004.
Øystese KA, Evang JA, Bollerslev J. Non-functioning pituitary adenomas: growth and aggressiveness. Endocrine. 2016;53(1):28–34. https://doi.org/10.1007/s12020-016-0940-7.
Yang Q, Li X. Molecular network basis of invasive pituitary adenoma: a review. Front Endocrinol. 2019;10:7. https://doi.org/10.3389/fendo.2019.00007.
Zhan X, Desiderio DM. Editorial: Systems biological aspects of pituitary tumors. Front Endocrinol. 2016;7:86. https://doi.org/10.3389/fendo.2016.00086.
Zhan X, Long Y. Exploration of molecular network variations in different subtypes of human nonfunctional pituitary adenomas. Front Endocrinol. 2016;7:13. https://doi.org/10.3389/fendo.2016.00013.
Zhan X, Long Y, Zhan X, Mu Y. Consideration of statistical vs. biological significances for omics data-based pathway network analysis. Med One. 2017;1:e170002. https://doi.org/10.20900/mo.20170002.
Seifirad S, Haghpanah V. Inappropriate modeling of chronic and complex disorders: how to reconsider the approach in the context of predictive, preventive and personalized medicine, and translational medicine. EPMA J. 2019;10(3):195–209. https://doi.org/10.1007/s13167-019-00176-z.
Janssens JP, Schuster K, Voss A. Preventive, predictive, and personalized medicine for effective and affordable cancer care. EPMA J. 2018;9(2):113–23. https://doi.org/10.1007/s13167-018-0130-1.
Zhan X, Desiderio DM, editors. Molecular network study of pituitary adenomas. Lausanne: Frontiers Media SA; 2020. ISBN: 978-2-88963-602-0. https://doi.org/10.3389/978-2-88963-602-0.
Banerjee S, Saxena N, Sengupta K, Banerjee SK. 17alpha-Estradiol-induced VEGF-A expression in rat pituitary tumor cells is mediated through ER independent but PI3K-Akt dependent signaling pathway. Biochem Biophys Res Commun. 2003;300(1):209–15. https://doi.org/10.1016/s0006-291x(02)02830-9.
Wang Z, Jiang C, Ganther H, Lü J. Antimitogenic and proapoptotic activities of methylseleninic acid in vascular endothelial cells and associated effects on PI3K-AKT, ERK, JNK and p38 MAPK signaling. Cancer Res. 2001;61(19):7171–8.
Smyth LM, Zhou Q, Nguyen B, Yu C, Lepisto EM, Arnedos M, et al. Characteristics and outcome of AKT1 E17K-mutant breast cancer defined through AACR Project GENIE, a clinicogenomic registry. Cancer Discov. 2020;10(4):526–35. https://doi.org/10.1158/2159-8290.CD-19-1209.
Iida M, Harari PM, Wheeler DL, Toulany M. Targeting AKT/PKB to improve treatment outcomes for solid tumors. Mutat Res. 2020;819-820:111690. https://doi.org/10.1016/j.mrfmmm.2020.111690.
Hunkeler M, Hagmann A, Stuttfeld E, Chami M, Guri Y, Stahlberg H, et al. Structural basis for regulation of human acetyl-CoA carboxylase. Nature. 2018;558(7710):470–4. https://doi.org/10.1038/s41586-018-0201-4.
Stoiber K, Nagło O, Pernpeintner C, Zhang S, Koeberle A, Ulrich M, et al. Targeting de novo lipogenesis as a novel approach in anti-cancer therapy. Br J Cancer. 2018;118(1):43–51. https://doi.org/10.1038/bjc.2017.374.
Fang W, Cui H, Yu D, Chen Y, Wang J, Yu G. Increased expression of phospho-acetyl-CoA carboxylase protein is an independent prognostic factor for human gastric cancer without lymph node metastasis. Med Oncol. 2014;31(7):15. https://doi.org/10.1007/s12032-014-0015-7.
Alkharusi A, Lesma E, Ancona S, Chiaramonte E, Nyström T, Gorio A, et al. Role of prolactin receptors in lymphangioleiomyomatosis. PLoS One. 2016;11(1):e0146653. https://doi.org/10.1371/journal.pone.0146653.
Zhao SJ, Jiang YQ, Xu NW, Li Q, Zhang Q, Wang SY, et al. SPARCL1 suppresses osteosarcoma metastasis and recruits macrophages by activation of canonical WNT/β-catenin signaling through stabilization of the WNT-receptor complex. Oncogene. 2018;37(8):1049–61. https://doi.org/10.1038/onc.2017.403.
Ma Y, Xu Y, Li L. SPARCL1 suppresses the proliferation and migration of human ovarian cancer cells via the MEK/ERK signaling. Exp Ther Med. 2018;16(4):3195–201. https://doi.org/10.3892/etm.2018.6575.
Aruleba RT, Adekiya TA, Oyinloye BE, Kappo AP. Structural studies of predicted ligand binding sites and molecular docking analysis of Slc2a4 as a therapeutic target for the treatment of cancer. Int J Mol Sci. 2018;19(2):386. https://doi.org/10.3390/ijms19020386.
Wang J, Ding N, Li Y, Cheng H, Wang D, Yang Q, et al. Insulin-like growth factor binding protein 5 (IGFBP5) functions as a tumor suppressor in human melanoma cells. Oncotarget. 2015;6(24):20636–49. https://doi.org/10.18632/oncotarget.4114.
Duan C, Allard JB. Insulin-like growth factor binding protein-5 in physiology and disease. Front Endocrinol. 2020;11:100. https://doi.org/10.3389/fendo.2020.00100.
Güllü G, Karabulut S, Akkiprik M. Functional roles and clinical values of insulin-like growth factor-binding protein-5 in different types of cancers. Chin J Cancer. 2012;31(6):266–80. https://doi.org/10.5732/cjc.011.10405.
Lloyd RV, Jin L. Analysis of chromogranin/secretogranin messenger RNAs in human pituitary adenomas. Diagn Mol Pathol. 1994;3(1):38–45. https://doi.org/10.1097/00019606-199403010-00007.
Lloyd RV, Jin L, Kulig E, Fields K. Molecular approaches for the analysis of chromogranins and secretogranins. Diagn Mol Pathol. 1992;1(1):2–15. https://doi.org/10.1097/00019606-199203000-00002.
Jin L, Chandler WF, Smart JB, England BG, Lloyd RV. Differentiation of human pituitary adenomas determines the pattern of chromogranin/secretogranin messenger ribonucleic acid expression. J Clin Endocrinol Metab. 1993;76(3):728–35. https://doi.org/10.1210/jcem.76.3.7680355.
d'Herbomez M, Do Cao C, Vezzosi D, Borzon-Chasot F, Baudin E, groupe des tumeurs endocrines (GTE France). Chromogranin A assay in clinical practice. Ann Endocrinol (Paris). 2010;71(4):274–80. https://doi.org/10.1016/j.ando.2010.04.004 Epub 2010 Jun 9.
Cruz-Topete D, Christensen B, Sackmann-Sala L, Okada S, Jorgensen JO, Kopchick JJ. Serum proteome changes in acromegalic patients following transsphenoidal surgery: novel biomarkers of disease activity. Eur J Endocrinol. 2011;164(2):157–67. https://doi.org/10.1530/EJE-10-0754.
Tang KT, Yang HJ, Choo KB, Lin HD, Fang SL, Braverman LE. A point mutation in the albumin gene in a Chinese patient with familial dysalbuminemic hyperthyroxinemia. Eur J Endocrinol. 1999;141(4):374–8. https://doi.org/10.1530/eje.0.1410374.
Liu X, Zheng W, Wang W, Shen H, Liu L, Lou W, et al. A new panel of pancreatic cancer biomarkers discovered using a mass spectrometry-based pipeline. Br J Cancer. 2017;117(12):1846–54. https://doi.org/10.1038/bjc.2017.365.
Lin X, Hong S, Huang J, Chen Y, Chen Y, Wu Z. Plasma apolipoprotein A1 levels at diagnosis are independent prognostic factors in invasive ductal breast cancer. Discov Med. 2017;23(127):247–58.
Hu CA, Klopfer EI, Ray PE. Human apolipoprotein L1 (ApoL1) in cancer and chronic kidney disease. FEBS Lett. 2012;586(7):947–55. https://doi.org/10.1016/j.febslet.2012.03.002.
Zhan X, Desiderio DM. Heterogeneity analysis of the human pituitary proteome. Clin Chem. 2003;49(10):1740–51. https://doi.org/10.1373/49.10.1740.
Moreno CS, Evans CO, Zhan X, Okor M, Desiderio DM, Oyesiku NM. Novel molecular signaling and classification of human clinically nonfunctional pituitary adenomas identified by gene expression profiling and proteomic analyses. Cancer Res. 2005;65(22):10214–22. https://doi.org/10.1158/0008-5472.CAN-05-0884.
Zhan X, Wang X, Long Y, Desiderio DM. Heterogeneity analysis of the proteomes in clinically nonfunctional pituitary adenomas. BMC Med Genet. 2014;7:69. https://doi.org/10.1186/s12920-014-0069-6.
Golubnitschaja O, Costigliola V, EPMA. General report & recommendations in predictive, preventive and personalised medicine 2012: white paper of the European Association for Predictive, Preventive and Personalised Medicine. EPMA J. 2012;3(1):14. https://doi.org/10.1186/1878-5085-3-14.
Hu R, Wang X, Zhan X. Multi-parameter systematic strategies for predictive, preventive and personalised medicine in cancer. EPMA J. 2013;4(1):2. https://doi.org/10.1186/1878-5085-4-2.
Acknowledgments
The authors also acknowledge Professor Xuejun Li from Xiangya Hospital and Professor Dominic M. Desiderio from University of Tennessee Health Science Center to assist in obtaining human tissues.
Funding
This work was supported by the Shandong First Medical University Talent Introduction Funds (to X.Z.), the Hunan Provincial Hundred Talent Plan (to X.Z.), the SCIBP supported project (No. SCIBP2019090006), and China “863” Plan Project (Grant No. 2014AA020610-1 to XZ).
Author information
Authors and Affiliations
Contributions
D.L. analyzed data, prepared figures and tables, and wrote the manuscript draft. J.J., N.L., M.L., and S.W. participated in partial data analysis and bioinformatics. X.Z. conceived the concept, designed and instructed the experiments, analyzed the data, obtained the phosphoproteomic data, supervised the results, coordinated, wrote and critically revised the manuscript, and was responsible for its financial supports and the corresponding works. All authors approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no conflict of interest.
Ethical approval
Four pituitary adenoma tissue samples, obtained from the Department of Neurosurgery of Xiangya Hospital, Central South University, were approved by the Xiangya Hospital Medical Ethics Committee of Central South University. Post-mortem control pituitary tissue samples, obtained from the Memphis Regional Medical Center (n = 5), were approved by the University of Tennessee Health Science Center Internal Review Board.
Additional information
Abbreviations for all particular genes and proteins can be found in the Supplemental Table 1 and the UniProtKB database at the following link: https://www.expasy.org/.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Liu, D., Li, J., Li, N. et al. Integration of quantitative phosphoproteomics and transcriptomics revealed phosphorylation-mediated molecular events as useful tools for a potential patient stratification and personalized treatment of human nonfunctional pituitary adenomas. EPMA Journal 11, 419–467 (2020). https://doi.org/10.1007/s13167-020-00215-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13167-020-00215-0
Keywords
- Nonfunctional pituitary adenomas
- Invasiveness
- Tandem mass tag (TMT) labeling
- TiO2 enrichment
- Quantitative phosphoproteomics
- Transcriptomics
- Differentially expressed genes
- Overlapped molecule (phosphoprotein
- invasive DEG)
- Signaling pathway
- Patient stratification
- Prognostic/predictive assessment
- Personalized treatment