Abstract
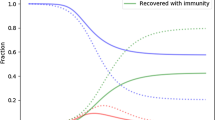
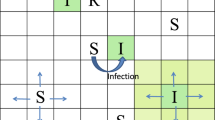
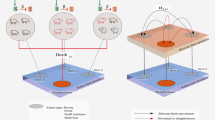
A network model at the population level and the individual level, which simulates both between-herd dynamics and within-herd dynamics, is formulated. We investigated effects of dispersal rates and local dynamics on the outcome of an epidemic at the population level. Numerical studies show that dispersal may strengthen spread of the disease on average, but lead to a less tendency for damped oscillation. Further, different types of clustering behaviors, from synchronized to completely desynchronized, are observed within our system. The results show that strengthening the coupling between farms via the immigration of infectives tends to enhance (in-phase) synchronization. Dynamic complexity, including chaotic, quasi-periodic or periodic behaviour, is observed in our model with varying the coupling strength. The main results help to explain differences in observed epidemiological patterns and to identify possible causes for different strains of Salmonella developing so much variation in their infection dynamics in UK dairy herds.








Similar content being viewed by others
References
Akkina, J.E., et al.: Epidemiologic aspects, control, and importance of multiple-drug resistant Salmonella typhimurium DT104 in the United States. J. Am. Vet. Med. Assoc. 214, 790–798 (1999)
Bergh, O., Getz, W.M.: Stability of discrete age-structured and aggregated delay-difference population models. J. Math. Biol. 26, 551–581 (1988)
Cooke, K., van den Driessche, P., Zou, X.: Interaction of maturation delay and nonlinear birth in population and epidemic models. J. Math. Biol. 39, 332–352 (1999)
Davies, R.H.: Salmonella typhimurium DT104:has it had its day? Practice 23, 342–351 (2001)
Davison, H.C., et al.: Prevalence, incidence and geographical distribution of serovars of Salmonella on dairy farms in England and Wales. Vet. Rec. 157, 703–711 (2005)
Diekmann, O., Heesterbeek, J.A.P., Metz, J.A.J.: On the definition and the computation of the basic reproduction ratio \(R_0\); in models for infectious diseases in heterogeneous populations. J. Math. Biol. 28, 365–382 (1990)
Earn, D.J.D., Rohani, P., Grenfell, B.T.: Persistence, chaos and synchrony in ecology and epidemiology. Proc. R. Soc. Lond. B 265, 7–10 (1998)
Ferguson, N.M., Donnelly, C.A., Anderson, R.M.: The foot-andmouth epidemic in Great Britain: pattern of spread and impact of interventions. Science 292, 1155–1160 (2001)
Fromont, E., Pontiera, D., Langlais, M.: Disease propagation in connected host populations with density-dependent dynamics: the case of the feline leukemia virus. J. Theor. Biol. 223, 465–475 (2003)
Gibbens, J.C., et al.: Descriptive epidemiology of the 2001 foot-and-mouth disease epidemic in Great Britain: the first five months. Vet. Rec. 149, 729–743 (2001)
Gilbert, M., Mitchell, A., Bourn, D., Mawdsley, J., Clifton- Hadley, R., Wint, W.: Cattle movements and bovine tuberculosis in Great Britain. Nature 435, 491–496 (2005)
Gotelli, N.J.: A primer of ecology. Sinauer, Sunderland (1998)
Grenfell, B.T., Bjonstad, O.N., Kappey, J.: Travelling waves and spatial hierarchies in measles epidemics. Nature 414, 716–723 (2001)
He, D., Stone, L.: Spatio-temporal synchronization of recurrent epidemics. Proc. R. Soc. Lond. Ser. B 270, 1519–1526 (2003)
Hethcote, H.W., van den Driessche, P.: Two SIS epidemiologic models with delays. J. Math. Biol. 240, 3–26 (2000)
Holland, M.D., Hastings, A.: Strong effect of dispersal network structure on ecological dynamics. Nature 456, 792–795 (2008)
Kaneko, K.: Clustering, coding, switching, hierarchical ordering, and control in network of chaotic elements. Physica D 41, 137–172 (1990)
Kaneko, K.: Relevance of dynamic clustering to biological networks. Physica D 75, 55–73 (1994)
Keeling, M.J.: The effects of local spatial structure on epidemiological invasions. Proc. R. Soc. Lond. Ser. B 266, 859–867 (1999)
Keeling, M.J., Rand, D.A., Morris, A.J.: Correlation models for childhood epidemics. Proc. R. Soc. B 264, 1149–1156 (1997)
Mainar-Jaime, R.C., Thurmond, M.C., Berzal-Herranz, B., Hietala, S.K.: Seroprevalence of Neospora caninum and abortion in dairy cows in northern Spain. Vet. Rec. 145, 72–75 (1999)
MAFF: Zoonoses Report United Kingdom 1999, 2000. Ministry of Agriculture. Fisheries and Food, UK (1999, 2000)
Newman, M.E.J., Watts, D.J.: Scaling and percolation in the small-world network model. Phys. Rev. E 60, 7332–7342 (1999)
Ruan, S., Wang, W., Levin, S.A.: The effect of global travel on the spread of SARS. Math. Biosci. Eng. 3, 205–218 (2006)
Schouten, J.M., Bouwknegt, M., van de Giessen, A.W., Frankena, K., De Jong, M.C.M., Graat, E.A.M.: Prevalence estimation and risk factors for Escherichia coli O157 on Dutch dairy farms. Prev. Vet. Med. 64, 49–61 (2004)
van den Driessche, P., Watmough, J.: Reproduction numbers and sub-threshold endemic equilibria for compartmental models of disease transmission. Math. Biosci. 180, 29–48 (2002)
van Schaik, G., Dijkhuizen, A.A., Huirne, R.B.M., Schukken, Y.H., Nielen, M., Hage, H.J.: Risk factors for existence of bovine herpes virus 1 antibodies on nonvaccinating Dutch dairy farms. Prev. Vet. Med. 34, 125–136 (1998)
Veterinary Laboratories Agency: Salmonella in Livestock Production in GB. VLA, Addlestore (1999, 2000, 2002)
Wang, W.D., Zhao, X.-Q.: An epidemic model in a patchy environment. Math. Biosci. 190, 97–112 (2004)
Wang, X., Xiao, Y., Wang, J., Lu, X.: A mathematical model of effects of environmental contamination and presence of volunteers on hospital infections in China. J. Theor. Biol. 293, 161–173 (2012)
Watts, D.J., Strogatz, S.H.: Collective dynamics of small-world networks. Nature 393, 440–442 (1998)
Wray, C., Davies, R.: Salmonella infections in cattle. In: Wray, C. (ed.) Salmonella in Domestic Animals, pp. 169–190. CABI Publishing, UK (2000)
Xiao, Y., Bowers, R.G., Clancy, D., French, N.P.: Understanding the dynamics of Salmonella infections in dairy herds: a modelling approach. J. Theor. Biol. 233, 159–175 (2005)
Xiao, Y., Clancy, D., French, N.P., Bowers, R.G.: A semi-stochastic model for Salmonella infection in a multigroup herd. Math. Biosci. 200, 214–233 (2006)
Xiao, Y., French, N., Bowers, R., Clancy, D.: Dynamics of infection with multiple transmission mechanisms in unmanaged/managed animal population. Theor. Popul. Biol. 71, 408–423 (2007)
Xiao, Y., French, N., Bowers, R., Clancy, D.: Pair approximations and the inclusion of indirect transmission: theory and application to between farm transmission Salmonella. J. Theor. Biol. 244, 532–540 (2007)
Xiao, Y., Zhou, Y., Tang, S.: Modelling disease spread in dispersal networks at two levels. IMA J. Math. Appl. Med. Biol. 28, 227–244 (2011)
Acknowledgments
The authors are supported by supported by the National Mega-project of Science Research (No. 2008ZX10001-003, YX), the National Natural Science Foundation of China NSFC (11171268 YX, 11571273 YX), and by the Fundamental Research Funds for the Central Universities (08143042 (YX)).
Author information
Authors and Affiliations
Corresponding author
Appendices
Appendix 1
1.1 Matrices contributing to the disease-free equilibrium \(E_0\)
Define a matrix M(z)
for any non-negative constant z. And matrix B gives
1.2 Matrices contributing to the next generation matrix H
where \( D_i =d_i +\alpha _i +\gamma _i \), \( i=1,\ldots , n\).
Appendix 2
The dispersal rates for the susceptibles, infectives and removeds are generated uniformly distributed to represent varying dispersal rates. In particular, we let \( h_S=2^{-2} rand(10,1)\); \(h_I= 2^{-2} rand(10,1); h_R= 2^{-2} rand(10,1)\). For Fig. 2b, the dispersal rates are \(h_b=(h_S, h_I, h_R)^T\), and
For Fig. 2c, the dispersal rates are \(h_c=(h_S, h_I, h_R)^T\), and
About this article
Cite this article
Wang, Y., Xiao, Y. An epidemic model on the dispersal networks at population and individual levels. Japan J. Indust. Appl. Math. 32, 641–659 (2015). https://doi.org/10.1007/s13160-015-0189-1
Received:
Revised:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13160-015-0189-1