Abstract
Ultra-conserved RNA (ucRNA) is a subset of long non-coding RNA, that is highly conserved among mice, rats and humans. UcRNA has attracted extensive attention in recent years for its potential biological significance in normal physiological function and diseases. However, due to the instability of RNA and the technical limitation, the function and mechanism of ucRNAs are largely unknown. Over the last two decades, researchers have made a lot of efforts to try to lift the veil of ucRNA in nervous, cardiovascular system and other systems as well as cancers. Since the concept of the glymphatic system is relatively new, we summarized here recent findings on the functions, regulation and the underlying mechanisms of ucRNAs in physiology and pathology. Meanwhile, pathology in some diseases is likely to contribute to abnormal expression of ucRNA in turn. We also discuss the technical challenges and bright prospects for future applications of ucRNAs in the diagnosis and treatment of diseases.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Ultra-conserved RNAs (ucRNAs) are transcribed from 481 conserved DNA fragments present in the genome. They represent a subset of long non-coding RNAs (lncRNAs), which are non-coding RNAs over 200 nucleotides in length. UcRNAs were first discovered in 2004 by Gill Bejerano et al., and are highly conserved among mice, rats, and humans [1].
UcRNAs are highly conserved not only among species, but also throughout evolution. The presence of evolutionary conservation indicates ucRNAs may have critical physiological functions. In addition, several studies have shown they play an important role in regulating gene expression during carcinogenesis and injury, which are considered to be low conserved or tissue-specific biological processes. However, many of the specific mechanisms underlying ucRNA function remain incompletely understood. Therefore, further research of ucRNAs is warranted to deepen understanding of the physiological and pathological significance of these molecules.
Past studies have demonstrated the expression levels of ucRNAs impact normal human development. Alterations in expression levels play a significant role in the development of various diseases, including heart defects, tuberculosis, and nerve pain, but also can be exploited to treat these disease states. In addition, studies have shown that many ucRNAs can play either a positive or negative regulatory role in cancer progression. For example, aberrantly expressed uc.63 + can act as a carcinogen to promote cancer cell invasion and migration in a variety of tumor tissues. Most research on ucRNAs thus far has focused on their important roles in cancer, and has uncovered their potential as diagnostic markers and therapeutic targets. This paper reviews the current scientific understanding of ucRNAs in human physiological development and pathological disease development, providing an important reference for further ucRNA research.
2 Biogenesis and functional mechanisms of ultra-conserved RNA
UcRNAs are transcripts from ultra-conserved regions (UCRs) which are widely transcribed across tissues. Of the 481 UCRs, 111 are designated as partly-exonics (E), which overlap with the known mRNAs of human protein-coding genes, thus exhibiting significant functions in RNA processing specificity. 256 non-exonics (N), which have no evidence of encoding proteins, play an important role in transcriptional regulation at the DNA level. For the remaining 114, the evidence for transcription is inconclusive and we called these possibly exonics (P).
Through microarray analysis, about 93% of the UCRs in the 19 specimens are expressed by background in at least one sample, we believe these as T-UCRs. The transcription frequencies of the three different types of UCRs (E, P, N) are similar. Of all the normal tissues analyzed, 84 of the 962 UCRs are transcribed bidirectionally, while 241 are transcribed from only one strand. In addition, strand-specific qRT-PCR and microarray analysis showed that about 83% of non-exonic T-UCRs are not intron transcripts of long precursor transcripts of known host genes, but truly independent non-coding transcripts [2].
Multiple expression techniques, including northern blot, qRT-PCR, and genome-wide microarray analysis, demonstrated that UCRs have distinct transcriptional signatures at vulnerable sites in the genome and cancer-associated genomic regions (CAGRs). This strong association between T-UCRs and predefined regions in the cancer genome suggests that these areas can be used as both therapeutic markers and targets [2]. T-UCR has been shown to drive malignant transformation of human diseases by regulating non-coding RNAs and/or protein-coding genes.
As a calss of long non-coding RNA, ucRNAs regulate target gene expression through multiple mechanisms. One proposed mechanism is that ucRNAs act as competitive endogenous RNAs (ceRNAs), which compete for binding to microRNAs (miRNAs), resulting in the stabilization of mRNA. In vitro, UCRs were cloned in luciferase reporter vectors to assess the possible direct interaction with miRNA, except that uc.348(N) did not interact with miR-155 as indicated by the luciferase assay. In vivo, the results were consistent. In addition, 87 miRNAs were found to be negatively correlated with T-UCRs expression levels [2]. Another proposed mechanism for ucRNA function is through the regulation of alternative splicing. Alternative splicing is a process by which different mRNA transcripts can be produced from the same gene, leading to the production of different protein isoforms. It has been suggested that ucRNAs may play a role in regulating alternative splicing by binding to splicing factors or other RNA-binding proteins [3]. UcRNAs may also function as scaffolds for the assembly of protein complexes involved in gene regulation [4]. For example, some ucRNAs have been shown to bind to both DNA and RNA-binding proteins, suggesting that they may play a role in the regulation of gene expression at the transcriptional or post-transcriptional level.
3 Physiology
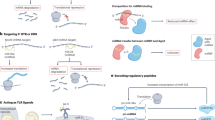
Normal physiological activities of biomolecules are critical for maintaining human health and development throughout life. Some ucRNAs have been found to influence physiological activities and the maintenance of physiological states (as shown in Fig. 1).
The roles and potential mechanisms of ucRNAs in physiological activities. Influences of each ucRNA are shown on the normal physiological activities of various systems and organs of the human body, mainly including heart, nerves, skin, kidney and intestine. At the same time, this figure also indicates the mechanisms of ucRNA to some extent
3.1 Neuroendocrine system
The nervous system controls the activities of various organs and systems, while the endocrine system is the most important metabolic regulator in the body. These body systems work together closely to modulate a wide variety of physiological functions. The expression of ucRNAs in these systems plays a critical role in both neuroendocrine development and regulation of neuroendocrine physiological activity.
It has been reported that the ucRNA, uc.189, promotes the development of the neocortex by limiting its expression to the ventricular zone (VZ) which is located in the nucleus. It was also identified the ucRNA, uc.189, can upregulate its regulatory target serine-arginine-rich splicing factor 3 (SRSF3) protein expression, which prevents the abnormal proliferation of neural progenitor cells [5]. E60UCS is important for brain development. It is transcribed from a 25 kb fragment downstream of the Pax6 3’ UTR in the large final intron of the adjacent Rlp4 gene. This ucRNA is mainly expressed in the optic cup, optic fold, telencephalon, diencephalon, and the precursor P3 peptide. E60UCS drives telencephalon enhancer activity by cooperating or hierarchizing with the Pax6 surrounding it and its expression remains unchanged throughout development [6]. The level of uc.173 was found to decrease significantly in the established mouse model representing lead induced nerve injury. They demonstrated uc.173 plays an anti-apoptotic role in lead induced nerve injury at multiple levels. MiR-291a-3p, a target gene of uc.173, has apoptosis-associated target genes (PI3K, NIK and Cn) and a DNA-damage repair-associated target gene (RAD23B). Therefore, they hypothesized that uc.173 may mediate apoptotic inhibition through miR-291a-3p, which has not been proved [7]. In another study, Wu found that uc.173 is highly upregulated in the prefrontal cortex of Sprague–Dawley (SD) rats after exposure to methylphenidate and may regulate neuronal differentiation of rat pheochromocytoma cells. They also found uc.173 and ubiquitin-conjugating enzyme E2B (UBE2B) have sensory overlap and thus participate in axonal regeneration of peripheral neurons [8]. Furthermore, it has been shown MeCP2 directly regulates uc.478 and uc.479 to influence the function of receptor and protein stability, thus decreasing GLUR-3 protein level, which induces the development of Rett syndrome (RTT) progressive neurodevelopmental disorder. Finally, Uc.309 expression was identified to be negatively correlated with the expression of an intron in Btrc, which modulates neural differentiation by degrading REST [9].
3.2 Cardiovascular system
The cardiovascular system is critical for the maintenance of a normal physiological state by supplying oxygen and nutrients to all cells in the body while also removing waste and carbon dioxide. Many studies have identified a role of ucRNAs in normal cardiovascular development and function.
A study found the ucRNA, T-uc.167, which is located on the antisense chain of the coding gene Mef2c, exhibits high expression in patients with ventricular septal defect (VSD). Uc.167 leads to G1 arrest while uc.40 prolongs S phase to reduce cell proliferation and differentiation and to induce apoptosis. In this mechanism, Mef2c is negatively regulated by uc.167, however, overexpression of Mef2c can reverse the effects of uc.167 [10]. Uc.40 also has a known role in inhibiting the proliferation and differentiation of cardiomyocytes by downregulating Pre-B-cell leukemia homeobox transcription factor 1 (PBX1) expression [11]. Interestingly, although uc.245 has no direct effects on the cell cycle, it can play the same role as the ucRNAs discussed above by downregulating the expression of friend of GATA 2 (FOG2). In addition, the co-silencing of uc.245 and FOG2 can upregulate the expression of specific genes and proteins required for myocardial formation, thus promoting the differentiation of P19 cells into cardiomyocytes [12]. Similar to uc.167, a study found that uc.4 is located in the Casz1 gene region but is expressed independently of this gene. Uc.4 does not affect cell proliferation and apoptosis, but its overexpression can inhibit the differentiation of P19 cells, which may have a negative impact on the development of cardiac structure and function [13].
3.3 Other systems
In lead induced renal tubular epithelial cells, it was found that the upregulation of uc.173 significantly promotes the viability of Human proximal tubular epithelial cell line (HK-2) and human kidney proximal tubular epithelial cell line (HKC) exposed to lead, and also restores the expression of apoptosis related genes such as Caspase-3, Caspase-9, Cleaved Caspase-3 and Cleaved Caspase-9. But the function of T-UCR in lead-induced nephrotoxicity is still unknown [14]. This ucRNA also has known roles in the function of the intestinal epithelial barrier. Uc.173 acts as a natural decoy RNA for miR-29b, which directly interacts with claudin-1 (CLDN1) mRNA via the 3' untranslated region and represses its translation. Normal expression of CLDN1 restores the intestinal epithelial barrier function [15]. In anouther study, it was found that uc.291 interacts with actin-like 6A (ACTL6A) to modulate chromatin remodeling in keratinocytes in the basal layer of the epidermis, therefore allowing the transcription of late differentiation genes of the epidermis. The decrease of uc.291 expression enhances the ability of ACTL6A to bind to the promoters of Brahma homologue/Brahma-related gene 1 (BRM/BRG1), and subsequently downregulates the expression of Edc. This mechanism inhibits the activation of epidermal differentiation genes. Therefore, uc.291 is important for the switch between the epidermal progenitor state and differentiation into mature epidermal cells [16].
4 Pathology
Disease is an extremely complex state in the body and can take many different forms. Interactions between pathogenic factors and the immune system can have dramatic impacts on cell and tissue morphology, metabolic states, and physiological function. Recent studies have identified potential roles of ucRNAs in the development of pathological disease states in various body systems(as listed in Table 1).
4.1 Neuroendocrine system
The development of neurological diseases is complex. The neurological impact of altered ucRNA expression caused by various diseases such as diabetes are summarized here. Diabetes may increase the expression of uc.360 + and uc.48 + and lead to neuropathy. Researchers found the silencing of these two ucRNAs could significantly reduce blood pressure, heart rate, and the ratio of low frequency to high frequency in heart rate variability, thus maintaining the dynamic balance between parasympathetic and sympathetic nerve activities [17, 18]. Mechanistically, they also found low expression of uc.360 + and uc.48 + can alleviate diabetic cardiac autonomic neuropathy (DCAN) and pain caused by diabetes and promote cardiac autonomic nerve remodeling by reducing the expression of P2X4, P2X3, and P2X7 respectively, thus resulting in the reduction of the level of inflammatory factors and inhibition of the ERK1/2 signaling pathway [18,19,20,21]. In addition, uc.48 + induces mechanical hyperalgesia and pain transmission of trigeminal neuralgia (TN) by increasing the expression of P2X7 [22]. Similarly, uc.48 + promotes human immunodeficiency virus (HIV) related neuropathic pain sensitivity by actively upregulating the P2Y12 receptor and downregulating p38 MAPK activity, thereby increasing intracellular cAMP concentration [23].
4.2 Cardiovascular system
A properly functioning heart is as critical to the body as a properly functioning engine is to a car. However, cardiac pathologies are common and can have detrimental impacts on the life of effected individuals. Currently, cardiovascular diseases are the leading cause of death worldwide. The ucRNA, uc.98, was found to be involved in regulating the physiological state of endothelial cells and the stability of plaque. Uc.98 was shown to result in abnormal proliferation of aortic endothelial cells (AEC), enhanced adhesion, and to significantly increase the levels of inflammatory transcription factors by activating phosphatidylinositol 3-kinase/Akt/nuclear factor (NF)-κB pathway and increasing the expressions of ICAM-1 and VCAM-1, thus accelerating the process of atherosclerosis (AS) [24]. The overexpression of uc.457 can inhibit the proliferation of P19 cells in a time-dependent manner and reduce the expression of genes related to myocardial maturation such as Histone Cell Cycle Regulator (HIRA), Natriuretic Peptide A (NPPA), Troponin T2, Cardiac Type (CTnT) and Myocyte Enhancer Factor 2C (Mef2c), which demonstrates aberrant uc.457 expression can induce coronary heart disease [25]. A study found uc.323 could prevent myocardial hypertrophy by inhibiting the expression of atrial natriuretic peptide (ANP) and B-type natriuretic peptide (BNP). In addition, uc.323 can form a negative feedback loop with mTORC1 to inhibit the phosphorylation of ERK1/2, form a complex with enhancer of zeste homolog 2 (EZH2) to inhibit the transcriptional expression of the Carnitine Palmitoyl transferase 1b (CPT1B) promoter, and regulate trimethylation of lysine 27 of histone H3 (H3K27me3) to inhibit cardiomyocyte hypertrophy [26].
4.3 Tumors
A tumor is an abnormal mass of tissue formed by local histiocytic hyperplasia under the action of various tumorigenic factors. Tumor development is a highly complex and multifactored process and can result in the formation of both benign or malignant tumors. Cancer results from the development of a malignant tumor which can spread both locally and systemically. Cancer is one of the leading causes of deaths worldwide and has no effective cure. A lot of studies have found that ucRNA plays an important role in cancer development (as listed in Table 2).
In the respiratory system, uc.338 can accelerate the migration and invasion of lung cancer cells by promoting Epithelial–mesenchymal transition (EMT) related proteins, in which CYCLIN B1, CDC25C, Snail, vimentin and N-cadherin are all upregulated [34]. In another study, uc.339 promotes the development and metastasis of lung adenocarcinoma in vivo by regulating the uc.339/miR-339/SLC7A11 axis, where the inhibition of miR-339 leads to increased expression of SLC7A11 and weakens ferroptosis [35]. In addition, although uc.83- is fully located in the Link01876 gene sequence, it can increase the number of cells in S-phase and promote the phosphorylation of p-AKT and p-ERK1 independently of Link01876 gene expression. Through this mechanism, uc.83- acts to promote the development of small cell lung cancer [36]. The ucRNA, uc.454, can negatively regulate its direct target gene Hspa12b to induce apoptosis of tumor cells, therefore acting as a tumor suppressor [37].
In esophageal carcinoma, uc.189 is transferred from esophageal squamous cell carcinomas (ESCC) to human lymphatic endothelial cells (HLECs) through exosomes, allowing ESCC cells to metastasize to distant locations through lymphatic vessels. In addition, uc.189 inhibits the translation of EPHA2 and increases the phosphorylation of P38MAPK and VEGF-C at the mRNA and protein levels, thereby promoting lymph angiogenesis and tumor lymphatic metastasis [38, 39].
In gastric cancer (GC), uc.160 + may inhibit the phosphorylation of AKT and the activation of MAPK signaling by regulating PTEN, thus inhibiting the proliferation, apoptosis, and viability of GC cells [40]. In another study, uc.160 + was found to bind directly with miR-155 to inhibit the growth of GC. Although there is a negative correlation between them, both act as tumor suppressors. This inconsistency warrants exploration into the role of uc.160 + in GC [41]. Conversely, overexpression of uc.63 + can induce p65 activity in the NF-κB complex leading to cell proliferation [42].
In the occurrence and development of hepatocellular carcinoma (HCC), the persistent low expression of uc.134, located in the intron region of Rsrc1, may lead to the low survival rate of HCC patients. Uc.134 can inhibit the development of HCC by inhibiting Cullin 4A (CUL4A) ubiquitination of Large Tumor Suppressor Kinase 1 (LATS1) and increasing YAPS127 phosphorylation, thereby activating the Hippo kinase signal [43]. In another study on liver cancer, it was found that the overexpression of uc.306 may lead to the further development of liver cancer. Uc.306 may be involved in EMT via BTRC (β-TrCP)-Wnt pathway, but the specific mechanism is not clear [44].
In recent years, the incidence rate of colorectal cancer has risen sharply. Therefore, studying the roles of ucRNAs in colorectal cancer is imperative. Uc.138 is located on chromosome 3 of Tra2b. It was identified that overexpression of uc.138 can increase cell proliferation by increasing G2 and S phases-related gene expression, including CDK1, cyclin A, and cyclin B and by downregulating genes related to cell death to reduce apoptosis. Consequently, this results in the resistance of colon cancer cells to anticancer drugs and promotes the migration of tumor cells [45]. In another study, uc.77- inhibited the growth of colorectal cancer (CRC) cells by blocking the G0/G1 phase through downregulation of the CDK4 protein (affected by E3 ligase). Interestingly, uc.77- can compete with F-Box and WD Repeat Domain Containing 8 (FBXW8) mRNA to bind to miR-4676-5p, thus promoting FBXW8 expression and ubiquitination of CDK4 to inhibit the proliferation of CRC cells [46].
In the urinary system, uc.416 + A, which is associated with renal cell carcinoma, and uc.8 + , which is associated with bladder cancer, negatively regulate their corresponding miRNAs to stimulate EMT and tumorigenesis [47, 48]. However, in contrast to uc.416 + A expression in renal cell carcinoma, the expression of uc.8 + is negatively correlated with the grade of bladder cancer by increasing the expression of matrix metalloproteinase 9 (MMP9). Another study on uc.8 + in the context of bladder cancer found it’s not only expressed in the early stage of bladder cancer, but also in the tumor microenvironment. The interaction network formed by DEAD-box helicase 19B (DDX19B) and nuclear RNA export factor 1 (NXF1) may participate in the transport of uc.8 + to the cytoplasm [49]. Uc.63 + induces androgen receptor (AR) expression in UMUC3 cells at the mRNA and protein levels, resulting in cisplatin (CDDP) resistance. Based on this finding, a study identified knockdown of uc.63 + can re-sensitize CDDP-resistant UMUC3 cells to drugs used for bladder cancer treatment [50].
Breast cancer (BC) is the most common cancer among women worldwide. The carcinogenic ucRNA, uc.147, resides in the 51st intron of the gene Lrba. Tacr3 and Cxxc4, which are positively correlated with their adjacent genes, were found to be related to estrogen receptors. Moreover, the combination of Cxxc4 and uc.147 can inhibit the expression of Cxxc4 and increase the invasiveness of tumors [51]. Uc.51 is also a carcinogenic ucRNA which acts at the transcriptional level by stably and positively regulating non-POU domain-containing octamer-binding protein (NONO) and increasing the phosphorylation of CREB [52]. Similarly, uc.63 can increase G2/M cells, inhibit cellular apoptosis, and promote BC and prostate cancer (PC) independently of the exportin-1 gene (Xpo1) [53]. Uc.63 + positively regulates AR, which leads to the resistance of prostate cancer cells to docetaxel. In addition, uc.63 + can also downregulate the expression of miR-130b of MMP2, thus playing a carcinogenic role [54]. Unlike the ucRNAs mentioned above, uc.38 is a tumor suppressor. The gene Pbx1 is negatively regulated as a downstream target gene of uc.38, which promotes apoptosis and inhibits proliferation by reducing BCL-2 expression and increasing BAX expression [55]. In the treatment of BC, Western blot analysis identified uc.57 can downregulate the expression of BCL11A at both the mRNA and protein level. It also acts to inhibit P13K/AKT and MAPK signaling pathways and subsequently reduce tamoxifen (TAM) resistance [56].
In central nervous system tumors, a study identified uc.283 + was overexpressed in gliomas while uc.160 + was expressed at a low level. However, the methylation of uc.160 + related CpG islands can promote the proapoptotic effect of RING1 and YY1-binding protein (RYBP) targets and forkhead box P2 (FOXP2) to regulate tumor suppressor genes and proto-oncogenes by regulating and editing pri-miR-376 clusters, thus leading to a better prognosis of glioma patients [57]. Conversely, uc.283 can negatively regulate ncRNA interaction to prevent pri-miRNA-195 from cleaving by Drosha [3].
Finally, ucRNAs also have identified roles in metastatic and fatal cutaneous squamous cell carcinoma (CSCC) derived skin cancer. The gene ACTL6A was found to regulate the SWI/SNF chromatin remodeling complex as a carcinogen in squamous cell carcinoma to maintain the tumor in an undifferentiated state. Interestingly, uc.291 can act negatively on ACTL6A, reversing the inhibition of differentiation genes mediated by ACTL6A [58].
5 Discussion
It is known that ultra-conserved RNA is conserved among humans, mice, and rats, which suggests ucRNAs are highly conserved among species. Not only ucRNAs are conserved among species, but they are also evolutionarily conserved indicating a critical developmental and/or physiological importance of these molecules. Therefore, research on ucRNAs has become increasingly prominent in recent years (as shown in Fig. 2). Despite increased research attention, ucRNA research is still in a stage of infancy compared to other biological molecules, such as DNA and proteins. Therefore, it is necessary to summarize the current understanding of ucRNAs and to prompt further research.
The publications related to ucRNAs in recent years. The number of papers related to ucRNA research is presented since the discovery of the ultra-conserved region in 2004. As can be seen from the figure, the number of papers generally maintains an upward trend, which means that ucRNA is attracting more and more attention and is worthy of further study
In terms of application prospects, ucRNA have the potential to be used as diagnostic markers and novel therapeutic targets for the treatment of various clinical diseases. For example, many ucRNAs can act as magnets to adsorb microRNAs, thus competing for binding sites with microRNAs. This results in negative interaction with the ucRNA’s corresponding microRNA and ultimately influences the occurrence and development of tumors. Similar mechanisms may also occur to facilitate the regulation of physiological and pathological changes in the human nervous system, digestive system, urinary system, and many other body systems. Mechanistically, ucRNAs also play a key role in the development of neuropathic pain by regulating ion channels and neuroinflammation. For example, activation of the P2X receptor by ucRNA promotes phosphorylation of ERK1/2 and the release of pro-inflammatory factors, which promote pain transmission and exacerbate mechanical and thermal hyperalgesia caused by various diseases. However, currently, the P2X receptor has only been shown to be involved in trigeminal neuralgia and pain caused by diabetes and acquired immunodeficiency syndrome (AIDS). Therefore, further study on the impact of this pathway in other types of pain, such as pain caused by cancer is warranted.
Although our understanding of the functions of ucRNA over the past two decades has dramatically increased, the application of ucRNA in disease treatment still faces many challenges. Thus, the development of ucRNA treatment methods is still in the pre-clinical or basic research phase. Translation from basic and clinical research to direct clinical applications that can alleviate disease burden requires increased research into the properties of these molecules. Compared with the stable properties of DNA and matured technologies associated with protein studies, RNA is easily degraded and damaged, and technological methods are unsatisfactory. In addition, much effort is still required to improve the quality of ucRNA research to more accurately and efficiently explore the specific intermolecular mechanisms of ucRNA functions. Future advancement in technology and research efforts will significantly expand the understanding of both the specific molecular signaling mechanisms of ucRNAs and the specific pedigree relationships with other RNAs and/or proteins. Using current knowledge and experience to study other RNAs in the lncRNA family, such as microRNAs, will provide a critical reference point to expand ucRNA research strategies. These studies will be invaluable for the developmental success of efficient and feasible protocols for ucRNAs to be implemented as clinical therapeutics.
Data availability
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
Code availability
Not applicable.
References
Bejerano G, Pheasant M, Makunin I, Stephen S, Kent WJ, Mattick JS, Haussler D. Ultraconserved elements in the human genome. Science. 2004. https://doi.org/10.1126/science.1098119.
Calin GA, Liu CG, Ferracin M, Hyslop T, Spizzo R, Sevignani C, Fabbri M, Cimmino A, Lee EJ, Wojcik SE, Shimizu M, Tili E, Rossi S, Taccioli C, Pichiorri F, Liu X, Zupo S, Herlea V, Gramantieri L, Lanza G, Alder H, Rassenti L, Volinia S, Schmittgen TD, Kipps TJ, Negrini M, Croce CM. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell. 2007. https://doi.org/10.1016/j.ccr.2007.07.027.
Galasso M, Dama P, Previati M, Sandhu S, Palatini J, Coppola V, Warner S, Sana ME, Zanella R, Abujarour R, Desponts C, Teitell MA, Garzon R, Calin G, Croce CM, Volinia S. A large scale expression study associates uc.283-plus lncRNA with pluripotent stem cells and human glioma. Genome Med. 2014. https://doi.org/10.1186/s13073-014-0076-4.
Taheri M, Ghafouri-Fard S. Long non-cod-ing RNA signature in cervical cancer. Klin Onkol. 2018. https://doi.org/10.4735/amko2018403.
Zhang M, Zhou J, Jiao L, Xu L, Hou L, Yin B, Qiang B, Lu S, Shu P, Peng X. Long non-coding RNA T-uc189 modulates neural progenitor cell fate by regulating srsf3 during mouse cerebral cortex development. Front Neurosci. 2021. https://doi.org/10.3389/fnins.2021.709684.
McBride DJ, Buckle A, van Heyningen V, Kleinjan DA. DNaseI hypersensitivity and ultraconservation reveal novel, interdependent long-range enhancers at the complex Pax6 cis-regulatory region. PLoS ONE. 2011. https://doi.org/10.1371/journal.pone.0028616.
Nan A, Zhou X, Chen L, Liu M, Zhang L, Luo Y, Liu Z, Dai L, Jiang Y. A transcribed ultraconserved noncoding RNA, Uc.173, is a key molecule for the inhabition of lead-induced neuronal apoptosis. Oncotarget. 2016;7(1):112–1243.
Wu T, Chen C, Yang L, Zhang M, Zhang X, Jia J, Wang J, Fu Z, Cui X, Ji C, Guo X, Tong M, Chen R, Hong Q, Chi X. Distinct lncRNA expression profiles in the prefrontal cortex of SD rats after exposure to methylphenidate. Biomed Pharmacother. 2015. https://doi.org/10.1016/j.biopha.2015.01.023.
Siqueira E, Obiols-Guardia A, Jorge-Torres OC, Oliveira-Mateos C, Soler M, Ramesh-Kumar D, Setien F, van Rossum D, Pascual-Alonso A, Xiol C, Ivan C, Shimizu M, Armstrong J, Calin GA, Pasterkamp RJ, Esteller M, Guil S. Analysis of the circRNA and T-UCR populations identifies convergent pathways in mouse and human models of Rett syndrome. Mol Ther Nucleic Acids. 2022. https://doi.org/10.1016/j.omtn.2021.12.030.
Song G, Shen Y, Ruan Z, Li X, Chen Y, Yuan W, Ding X, Zhu L, Qian L. LncRNA-uc.167 influences cell proliferation, apoptosis and differentiation of P19 cells by regulating Mef2c. Gene. 2016. https://doi.org/10.1016/j.gene.2016.06.006.
Wu R, Xue P, Wan Y, Wang S, Gu M. LncRNA-uc.40 silence promotes P19 embryonic cells differentiation to cardiomyocyte via the PBX1 gene. In Vitro Cell Dev Biol Anim. 2018. https://doi.org/10.1007/s11626-018-0284-0.
Liu H, Hu Y, Yin J, Yan X, Chen W, Wang X, Han S, Yu Z, Li M. Effects of long non-coding RNA uc.245 on cardiomyocyte-like differentiation in P19 cells via FOG2. Gene. 2019. https://doi.org/10.1016/j.gene.2018.12.080.
Cheng Z, Zhang Q, Yin A, Feng M, Li H, Liu H, Li Y, Qian L. The long non-coding RNA uc.4 influences cell differentiation through the TGF-beta signaling pathway. Exp Mol Med. 2018. https://doi.org/10.1038/emm.2017.278.
Qin J, Ning H, Zhou Y, Hu Y, Huang B, Wu Y, Huang R. LncRNA Uc.173 is a key molecule for the regulation of lead-induced renal tubular epithelial cell apoptosis. Biomed Pharmacother. 2018;100:101–7. https://doi.org/10.1016/j.biopha.2018.01.112.
Wang JY, Cui YH, Xiao L, Chung HK, Zhang Y, Rao JN, Gorospe M, Wang JY. Regulation of intestinal epithelial barrier function by long noncoding RNA uc.173 through interaction with microRNA 29b. Mol Cell Biol. 2018. https://doi.org/10.1128/MCB.00010-18.
Panatta E, Lena AM, Mancini M, Smirnov A, Marini A, Delli Ponti R, Botta-Orfila T, Tartaglia GG, Mauriello A, Zhang X, Calin GA, Melino G, Candi E. Long non-coding RNA uc.291 controls epithelial differentiation by interfering with the ACTL6A/BAF complex. EMBO Rep. 2020. https://doi.org/10.5252/embr.201846734.
Wu B, Zhang C, Zou L, Ma Y, Huang K, Lv Q, Zhang X, Wang S, Xue Y, Yi Z, Jia T, Zhao S, Liu S, Xu H, Li G, Liang S. LncRNA uc.48+ siRNA improved diabetic sympathetic neuropathy in type 2 diabetic rats mediated by P2X7 receptor in SCG. Auton Neurosci. 2016. https://doi.org/10.1016/j.autneu.2016.04.001.
Wu H, Jiang M, Liu Q, Wen F, Nie Y. lncRNA uc48+ regulates immune and inflammatory reactions mediated by the P2X7 receptor in type 2 diabetic mice. Exp Ther Med. 2020. https://doi.org/10.3892/etm.2020.9360.
Wang S, Xu H, Zou L, Xie J, Wu H, Wu B, Yi Z, Lv Q, Zhang X, Ying M, Liu S, Li G, Gao Y, Xu C, Zhang C, Xue Y, Liang S. LncRNA uc48+ is involved in diabetic neuropathic pain mediated by the P2X3 receptor in the dorsal root ganglia. Purinergic Signal. 2016. https://doi.org/10.1007/s11302-015-9488-x.
Wu H, Wen F, Jiang M, Liu Q, Nie Y. LncRNA uc.48+ is involved in the diabetic immune and inflammatory responses mediated by P2X7 receptor in RAW2647 macrophages. Int J Mol Med. 2018. https://doi.org/10.3892/ijmm.2018.3661.
Xiong W, Huang L, Shen Y, Guan S, He L, Tong Z, Tan M, Liu L, Gao Y. Effects of lncRNA uc.48+ siRNA on the release of CGRP in the spinal cords of rats with diabetic neuropathic pain. Int J Clin Exp Pathol. 2017;10(9):9960–99693.
Xiong W, Tan M, Tong Z, Yin C, He L, Liu L, Shen Y, Guan S, Ge H, Li G, Liang S, Gao Y. Effects of long non-coding RNA uc.48+ on pain transmission in trigeminal neuralgia. Brain Research Bulletin. 2019. https://doi.org/10.1016/j.brainresbull.2019.02.009.
Peng L, Wu B, Shi L, Zou L, Li L, Yang R, Xu X, Li G, Liu S, Zhang C, Liang S. Long non-coding RNA Uc.48+ small interfering RNA alleviates neuroinflammatory hyperalgesia in Gp120-treated rats via the P2Y12 receptor. Front Neurosci. 2021. https://doi.org/10.3389/fnins.2021.663962.
Fan Z, Zhang Y, Xiao D, Ma J, Liu H, Shen L, Zhang M, He B. Long noncoding RNA UC.98 stabilizes atherosclerotic plaques by promoting the proliferation and adhesive capacity in murine aortic endothelial cells. Acta Biochim Biophys Sin. 2020. https://doi.org/10.1093/abbs/gmz155.
Zhang Q, Cheng Z, Yu Z, Zhu C, Qian L. Role of lncRNA uc.457 in the differentiation and maturation of cardiomyocytes. Mol Med Rep. 2019. https://doi.org/10.3892/mmr.2019.10132.
Sun Y, Fan W, Xue R, Dong B, Liang Z, Chen C, Li J, Wang Y, Zhao J, Huang H, Jiang J, Wu Z, Dai G, Fang R, Yan Y, Yang T, Huang ZP, Dong Y, Liu C. Transcribed ultraconserved regions, Uc.323, ameliorates cardiac hypertrophy by regulating the transcription of CPT1b (carnitine palmitoyl transferase 1b). Hypertension. 2020. https://doi.org/10.1161/HYPERTENSIONAHA.119.13173.
Shi L, Sun M, Ren X, Li Z, Yang R, Xu X, Li L, Li G, Liu S, Schmalzing G, Nie H, Li G, Liang S. LncRNA UC.360+ shRNA improves diabetic cardiac sympathetic dysfunction mediated by the P2X4 receptor in the stellate ganglion. ACS Chem Neurosci. 2021. https://doi.org/10.1021/acschemneuro.1c00050.
Li ZB, Han YS, Wei LL, Shi LY, Yi WJ, Chen J, Huang H, Jiang TT, Li JC. Screening and identification of plasma lncRNAs uc.48+ and NR_105053 as potential novel biomarkers for cured pulmonary tuberculosis. Int J Infect Dis. 2020. https://doi.org/10.1016/j.ijid.2020.01.005.
Guo J, Fang W, Sun L, Lu Y, Dou L, Huang X, Tang W, Yu L, Li J. Ultraconserved element uc.372 drives hepatic lipid accumulation by suppressing miR-195/miR4668 maturation. Nat Commun. 2018. https://doi.org/10.1038/s41467-018-03072-8.
Zhang Y, Sun J, Yao H, Lin Y, Wei J, Hu G, Guo J, Li J. Ultraconserved element uc.333 increases insulin sensitivity by binding to miR-223. Aging. 2020. https://doi.org/10.18632/aging.103020.
Qian XX, Peng JC, Xu AT, Zhao D, Qiao YQ, Wang TR, Shen J, Ran ZH. Noncoding transcribed ultraconserved region (T-UCR) uc.261 participates in intestinal mucosa barrier damage in crohn’s disease. Inflamm Bowel Dis. 2016. https://doi.org/10.1097/MIB.0000000000000945.
Qian XX, Cai CW, Li HY, Lai LJ, Song DJ, Qiao YQ, Shen J, Ran ZH. Transcribed ultraconserved region (T-UCR) uc.261 expression is closely correlated with disease activity and intestinal permeability in Crohn’s disease. Therap Adv Gastroenterol. 2019. https://doi.org/10.1177/1756284819880733.
Yu M, Guan Z, Li S, Wen X, Shi H, Qu G, Lu X, Zhu X, Wang B, Feng Q, Gan W, Zhang A. Gene expression profiling analysis reveals that the long noncoding RNA uc.412 is involved in mesangial cell proliferation. Mol Med Rep. 2019. https://doi.org/10.3892/mmr.2019.10753.
Gao X, Gao X, Li C, Zhang Y, Gao L. Knockdown of long noncoding RNA uc338 by siRNA inhibits cellular migration and invasion in human lung cancer cells. Oncol Res. 2016. https://doi.org/10.3727/096504016x14666990347671.
Zhang N, Huang J, Xu M, Wang Y. LncRNA T-UCR Uc.339/miR-339/SLC7A11 axis regulates the metastasis of ferroptosis-induced lung adenocarcinoma. J Cancer. 2022. https://doi.org/10.7150/jca.65017.
Vannini I, Ferracin M, Fabbri F, Fabbri M. Overexpression of ultraconserved region 83- induces lung cancer tumorigenesis. PLoS ONE. 2022. https://doi.org/10.1371/journal.pone.0261464.
Zhou J, Wang C, Gong W, Wu Y, Xue H, Jiang Z, Shi M. Inhibited growth by targeting heat shock protein family a member 12b in non-small-cell lung cancer. Mol Ther Nucleic Acids. 2018. https://doi.org/10.1016/j.omtn.2018.05.004.
Guo Y, Wang C, Miao X, Chen S, Qian Y, Li G, Jiang Y. Upregulation of uc.189 in patients with esophageal squamous cell carcinoma and its clinicopathologic value. Pathol Res Pract. 2017. https://doi.org/10.1016/j.prp.2017.09.005.
Ding Z, Yan Y, Guo YL, Wang C. Esophageal carcinoma cell-excreted exosomal uc189 promotes lymphatic metastasis. Aging. 2021. https://doi.org/10.18632/aging.202979.
Honma R, Goto K, Sakamoto N, Sekino Y, Sentani K, Oue N, Yasui W. Expression and function of Uc.160+, a transcribed ultraconserved region, in gastric cancer. Gastric Cancer. 2017. https://doi.org/10.1007/s10120-017-0714-9.
Pang L, Li Q, Zhang Y, Deng B, Wu F, Wang J, Wu K, Ding Y, Yu D. Transcribed ultraconserved noncoding RNA uc.160 acts as a negative regulator in gastric cancer. Am J Transl Res. 2018;10(9):2822–33.
Sakamoto N, Sekino Y, Fukada K, Pham QT, Honma R, Taniyama D, Ukai S, Takashima T, Hattori T, Naka K, Tanabe K, Ohdan H, Yasui W. Uc.63+ contributes to gastric cancer progression through regulation of NF-kB signaling. Gastric Cancer. 2020;23(5):863–73. https://doi.org/10.1007/s10120-020-01070-8.
Ni W, Zhang Y, Zhan Z, Ye F, Liang Y, Huang J, Chen K, Chen L, Ding Y. A novel lncRNA uc.134 represses hepatocellular carcinoma progression by inhibiting CUL4A-mediated ubiquitination of LATS1. J Hematol Oncol. 2017. https://doi.org/10.1186/s13045-017-0449-4.
Luo HL, Chen J, Luo T, Wu FX, Liu JJ, Wang HF, Chen M, Li LQ, Li H. Downregulation of macrophage-derived T-UCR uc.306 associates with poor prognosis in hepatocellular carcinoma. Cell Physiol Biochem. 2017. https://doi.org/10.1159/000479269.
Kuwano Y, Nishida K, Rokutan K. Overexpression of the transcribed ultraconserved region Uc.138 accelerates colon cancer progression. Sci Rep. 2021. https://doi.org/10.1038/s41598-021-88123-9.
Zheng Z, Hong D, Zhang X, Chang Y, Sun N, Lin Z, Li H, Huang S, Zhang R, Xie Q, Huang H, Jin H. Downregulation promotes colorectal cancer cell proliferation by inhibiting FBXW8-Mediated CDK4 protein degradation. Front Oncol. 2021. https://doi.org/10.3389/fonc.2021.673223.
Sekino Y, Sakamoto N, Goto K, Honma R, Shigematsu Y, Quoc TP, Sentani K, Oue N, Teishima J, Kawakami F, Karam JA, Sircar K, Matsubara A, Yasui W. Uc.416 + a promotes epithelial-to-mesenchymal transition through miR-153 in renal cell carcinoma. BMC Cancer. 2018. https://doi.org/10.1186/s12885-018-4863-y.
Olivieri M, Ferro M, Terreri S, Durso M, Romanelli A, Avitabile C, De Cobelli O, Messere A, Bruzzese D, Vannini I, Marinelli L, Novellino E, Zhang W, Incoronato M, Ilardi G, Staibano S, Marra L, Franco R, Perdonà S, Terracciano D, Czerniak B, Liguori GL, Colonna V, Fabbri M, Febbraio F, Calin GA, Cimmino A. Long non-coding RNA containing ultraconserved genomic region 8 promotes bladder cancer tumorigenesis. Oncotarget. 2016. https://doi.org/10.18632/oncotarget.7833.
Terreri S, Mancinelli S, Ferro M, Vitale MC, Perdona S, Castaldo L, Gigantino V, Mercadante V, De Cecio R, Aquino G, Montella M, Angelini C, Del Prete E, Aprile M, Ciaramella A, Liguori GL, Costa V, Calin GA, La Civita E, Terracciano D, Febbraio F, Cimmino A. Subcellular localization of uc.8+ as a prognostic biomarker in bladder cancer tissue. Cancers. 2021. https://doi.org/10.3390/cancers13040681.
Sekino Y, Sakamoto N, Ishikawa A, Honma R, Shigematsu Y, Hayashi T, Sentani K, Oue N, Teishima J, Matsubara A, Yasui W. Transcribed ultraconserved region Uc.63+ promotes resistance to cisplatin through regulation of androgen receptor signaling in bladder cancer. Oncol Rep. 2019. https://doi.org/10.3892/or.2019.7039.
Pereira Zambalde E, Bayraktar R, Schultz Jucoski T, Ivan C, Rodrigues AC, Mathias C, Knutsen E, Silveira de Lima R, Fiori Gradia D, de Souza Fonseca Ribeiro EM, Hannash S, Adrian Calin G, Carvalhode OJ. A novel lncRNA derived from an ultraconserved region: lnc-uc.147, a potential biomarker in luminal a breast cancer. RNA Biol. 2021. https://doi.org/10.1080/15476286.2021.1952757.
Shi X, Huang X, Chen R, Li Y, Xu Y, Zhang W, Zhu Q, Zha X, Wang J. The transcribed ultraconserved element uc.51 promotes the proliferation and metastasis of breast cancer by stabilizing NONO. Clin Exp Metastasis. 2021. https://doi.org/10.1007/s10585-021-10128-5.
Marini A, Lena AM, Panatta E, Ivan C, Han L, Liang H, Annicchiarico-Petruzzelli M, Di Daniele N, Calin GA, Candi E, Melino G. Ultraconserved long non-coding RNA uc.63 in breast cancer. Oncotarget. 2017. https://doi.org/10.18632/oncotarget.10572.
Sekino Y, Sakamoto N, Goto K, Honma R, Shigematsu Y, Sentani K, Oue N, Teishima J, Matsubara A, Yasui W. Transcribed ultraconserved region Uc.63+ promotes resistance to docetaxel through regulation of androgen receptor signaling in prostate cancer. Oncotarget. 2017. https://doi.org/10.18632/oncotarget.21688.
Zhang LX, Xu L, Zhang CH, Lu YH, Ji TH, Ling LJ. Induces breast cancer cell apoptosis via PBX1. Am J Cancer Res. 2017;7(12):2438–24513.
Zhang CH, Wang J, Zhang LX, Lu YH, Ji TH, Xu L, Ling LJ. Shikonin reduces tamoxifen resistance through long non-coding RNA uc.57. Oncotarget. 2017. https://doi.org/10.18632/oncotarget.20809.
Soler M, Davalos V, Sanchez-Castillo A, Mora-Martinez C, Setien F, Siqueira E, Castro de Moura M, Esteller M, Guil S. The transcribed ultraconserved region uc.160+ enhances processing and A-to-I editing of the miR-376 cluster: hypermethylation improves glioma prognosis. Mol Oncol. 2022. https://doi.org/10.1002/1878-0261.13121.
Mancini M, Cappello A, Pecorari R, Lena AM, Montanaro M, Fania L, Ricci F, Di Lella G, Piro MC, Abeni D, Dellambra E, Mauriello A, Melino G, Candi E. Involvement of transcribed lncRNA uc.291 and SWI/SNF complex in cutaneous squamous cell carcinoma. Discov Oncol. 2021. https://doi.org/10.1007/s12672-021-00409-6.
Acknowledgements
We thank Ashley Kuehnl from University of Wisconsin-Madison for helping with the English polish.
Funding
This work was supported by the National Natural Science Foundation of China (No. 81900194 to ZL) and the Start-up Foundation of Yangzhou University to ZL.
Author information
Authors and Affiliations
Contributions
Tingye Wang and Zhanping Lu conceived and drafted the manuscript. Tingye Wang and Feng Li prepared the graphs. Zhanping Lu finalized the manuscript. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
No potential competing interest was reported by the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wang, T., Li, F. & Lu, Z. Ultra-conserved RNA: a novel biological tool with diagnostic and therapeutic potential. Discov Onc 14, 41 (2023). https://doi.org/10.1007/s12672-023-00650-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12672-023-00650-1