Abstract
Gastric cancer (GC) has a great fatality rate, meanwhile, there is still a lack of available biomarkers for prognosis. The goal of the research was to discover key and novel potential biomarkers for GC. We screened for the expression of significantly altered genes based on survival rates from two consensus molecular subtypes (CMS) of GC. Subsequently, functional enrichment analysis showed these genes involved in many cancers. And we picked 6 hub genes that could both secreted in the tumor microenvironment and expression enhanced in immune cells. Then, Kaplan Meier survival and expression detected in the tumor pathological stage were utilized to clarify the prognostic of these 6 hub genes. The results indicated that OGN, CHRDL2, C2orf40, THBS4, CHRDL1, and ANGPTL1, respectively, were significantly associated with poor OS in GC patients. And their expression increased with cancer advanced. Moreover, immune infiltration analysis displayed that those hub genes expression positively with M2 macrophage, CD8+ T Cell, most immune inhibitors, and majority immunostimulators. In summary, our results suggested that OGN, CHRDL2, C2orf40, THBS4, CHRDL1, and ANGPTL1 were all potential biomarkers for GC prognosis and might also be potential therapeutic targets for GC.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Gastric cancer (GC) is one of the most common malignant tumors, and its mortality ranks fourth among all malignant tumors in the world [1]. According to the global cancer statistics (2020), there were over one million new cases of GC and with a mortality rate of 7.7% of the total cancer deaths [1]. Due to the lack of effective biomarkers, most patients with GC are diagnosed at an advanced stage and distant metastasis often occurs at this stage, leading to a poor prognosis. The 5-year overall survival rate of GC is about 20–30% in most parts of the world [2]. The earlier the effective intervention for GC patients, the more beneficial to improve their survival rate. Therefore, it is necessary to identify the key prognostic biomarkers in early time for GC therapy.
Immune checkpoint blockade (ICB) has been brought new hope for many cancers treatment. While ICB does not valid in most cases, including GC [3], and thus more research is required to drive the field forward. Immune-related cell in the tumor microenvironment (TME) is also an indispensable factor for the development, progression, and prognosis of GC. Despite CD8+ T cells are considered to be an anti-tumor component in the TME [4], studies on the relationship between CD8+ T cells and prognosis in GC yielded in divergence [5, 6]. Programmed cell death protein 1 (PD-1) is an important immune checkpoint molecule in CD8+ T cell and its upregulation disturbs the anti-tumor function of CD8+ T cell [7]. PD-1+CD8+ T cell infiltration was significantly increased in patients with GC [6]. While PD-1 blockade combined with immune modulators, such as TGF-β1 inhibitor, might take patients more benefits than targeted PD-1 alone [6]. And it was also reported that some subsets of CD8+ T, such as CXCR5+CD8+ T [8], but not CD8+ T itself connected with better overall survival (OS) in patients with GC. Therefore, it is important to find out the key molecules that regulate immune modulators in TME or/and co-express molecular in CD8+ T that mediated its function.
Besides, the latest studies have proved that macrophages, mostly M2 polarized macrophages, have a vital role in the GC advance [9, 10]. Infiltration of M2 macrophage might be a worthwhile biomarker for GC prognosis [11]. Targeting M2 macrophages may be a promising therapeutic strategy, therefore, it is valuable that uncover M2-related molecules.
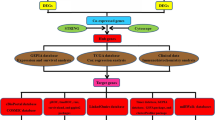
Here, we picked out those genes that were significantly associated with the survival of GC patients. And selected hub genes, OGN, CHRDL2, C2orf40, THBS4, CHRDL1, and ANGPTL1, which secreted into TME and enhanced in immune cells. After validated their expression value for GC diagnosis, we performed immune infiltration analysis of these 6 hub genes. Our study might provide novel biomarkers for GC advance, and potential targets that combined using with ICB to improve GC therapy.
2 Results
2.1 Identification of robust genes associated with GC survival
The COMSUC web server [12] was used to find out the hub genes which significantly correlated with the survival of GC patients. We firstly clustered GC samples into two consensus molecular subtypes (CMS) by multiple methods, including K-means, hierarchical clustering (Hclust), non-negative matrix factorization (NMF) (Supplementary Fig. S1a, b). Among them, we found that 52 genes prominently up-regulated expression in CMS1 compare to CMS2 (Fig. 1a). And CMS1 had lower survival than CMS2 group (Fig. 1b). These genes may act independently or work in concert for GC survival.
Identification of robust genes associated with GC survival and their functional enrichment analysis. a Identification of key modules that prominent change in CMS1 vs. CMS2. b Kaplan–Meier survival analysis showed CMS1 had poor survival. c Gene ontology (GO) enrichment analysis of CMS selected genes in Metascape
2.2 Functional enrichment analysis of survival-related CMS genes
To identify the notably enriched pathway and biological process of survival-related CMS genes, we explored in Metascape website [13] and found those genes related to many processes of cancer (Fig. 1c). And the most relative pathway was early gastric cancer (Fig. 1c). Then we analyzed the sub-network of those CMS1 genes in TCGA RNAseq of STAD, most of them had some connections in each other (Supplementary Fig. S1c). According to those results, we speculated whether those genes were regulated by some common transcription factors (TFs). After performing TF enrichment in Knock TF (http://www.licpathway.net/KnockTF/index.html), more than ten TFs predicted may regulate over 73% of genes of these selected 52 genes (Table 1). The top 4 TFs were TFAP4, TP53, CREB1, and PTEN, respectively (Supplementary Fig. S1d), and they all have been reported to promote the progression of GC [14,15,16,17]. Furthermore, we explored the characteristics of the 52 genes in the GeneCards (https://www.genecards.org/) and the Human Protein Atlas (https://www.proteinatlas.org/). As shown in Table 2, 10 genes could be transcribed into secreted proteins, 22 genes enriched in immune cells and only 7 genes own both characters (Supplementary Fig. S1e).
2.3 Survival outcomes in selected genes
To verify whether the selected 7 genes (OGN, CHRDL2, C2orf40, THBS4, CHRDL1, ANGPTL1, SLIT2) were correlated with worse overall survival (OS) in GC patients, survival analysis was performed in Kaplan Meier plotter [18]. On account of many articles that have been shown that SLIT2 was over-expression in GC and might be an independent risk factor for GC [19, 20], we did not explore it further in this paper. The Kaplan Meier plotter results showed that the up-regulated expression of OGN, CHRDL2, C2orf40, THBS4, CHRDL1, and ANGPTL1, respectively, was correlated with poor OS in GC patients (Fig. 2a–f). We then observed the prognostic value of these six hub genes in OncoLnc websites [21] and got similar results (Fig. 3a–f). Among them, OGN and CHRDL2 were firstly reported in GC that their expression was related to OS of GC patients. To further confirm the correlation of 6 genes expression with tumor progression in GC, expression in different stages was executed in GEPIA [22]. The analyzed results indicated that the expression of OGN, CHRDL2, C2orf40, THBS4, CHRDL1, and ANGPTL1 was gradually increased in the GC stage (Supplementary Fig. S2a–f). These results revealed these 6 hub genes were significantly related to the aggravation and survival of GC.
2.4 Immune cell infiltration analysis
To investigate the relationship between the expression of these hub genes (OGN, CHRDL2, C2orf40, THBS4, CHRDL1, and ANGPTL1) and immune infiltration, the TIMER website [23] was used to analyze the correlation between these hub genes and multiple immune cells. The results indicated that the expression of these hub genes was significantly positively correlated with CD8+ T cells, CD4+ T cells, macrophages, neutrophils, dendritic cells (Supplementary Fig. S3). Next, we further analyzed the correlations of macrophage subtypes infiltration with these hub genes in GC. The results exhibited these 6 hub genes had no significant link in M1-related markers with their expression, but exhibited remarkable connection in M2 and TAMs (Fig. 4a–c). We then picked OGN to experimentally validate its expression and its relationship with M2 macrophage. The expression of OGN was increased with GC advance (Fig. 5a), which was consistent with its mRNA expression level showed in GEPIA (Supplementary Fig. S2a). Moreover, our results showed OGN expression correlated with infiltrating of M2 macrophage in different stages of GC (Fig. 5a–c). The above results indicated that these 6 hub genes were associated with immune cell infiltration, while the relationship still needs more evidence to prove.
Expression of OGN and its relationship with M2 macrophage in different stages of GC. a Western blot showed the expression of OGN and CD163 (marker of M2 macrophage) in different stages of GC. b Correlation analysis verified OGN was positively related with CD163 in GC. c Representative images of OGN and CD163 in different stages of GC detected by immunohistochemistry staining
2.5 Correlation analysis between hub genes and immune markers
To better understand the immune function of these hub genes, we explored the relationship between their expression and immune markers in TISIDB [24]. Tumor immunology analysis exhibited that the expression of these 6 genes was positively correlated with most immunoinhibitors (Fig. 6a) and majority immunostimulators (Supplementary Fig. S4a). Furthermore, we check out the striking different genes verified again in GEPIA, like CSF1R and Cxcl12, and got similar results (Fig. 6b–g, Supplementary Fig. S4b–g). In the end, we search data found that the expression of OGN, C2orf40, CHRDL2, THBS4, CHRDL1, and ANGPTL1, respectively, correlated with different immune subtypes in GC (Fig. 7a–f). Molecular classification of GC benefited patients to gain precisely targeted therapies [25]. Previous studies that separately GC patients into 5 subtypes containing Epstein-Barr virus (EBV)-positive, hypermutated-single-nucleotide variant predominant (HM-SNV), hypermutated enriched for insertion/deletion (HM-indel, which lead to microsatellite instability), chromosomal instability (CIN), and genomically stable (GS) [25, 26]. We then explored the six hub genes expression in various molecular subtypes in GC (Supplementary Fig. S5a–f). In summary, these results implied that these 6 hub genes might potentially govern the conscription and awakening of immune cells in GC.
3 Discussion
Chemotherapy, radiotherapy, surgery, immunotherapy, and targeted therapy are effective in the progression of GC in appropriate indications. While the limited OS that no more than 30% in most countries, there are still need specific biomarkers to guide rational using drugs, especially for ICB treatment.
OGN is a member of the small leucine-rich proteoglycan (SLRP) family and its function may vary from different tumors. However, this is the first time that the role of OGN in GC has been reported so far. There was experimental evidence that OGN was upregulated and function as a tumor promoter by inhibiting NF2 expression and triggering mTOR signaling in meningioma [27], but OGN expression reduced and inhibited cell proliferation, invasion, and epithelial to mesenchymal transition through EGFR/Akt pathway in colorectal cancers [28]. While the work model of OGN in GC is not clear and needs to be further explored. Moreover, OGN, as one of the biologically active elements of the vascular extracellular matrix, could be tested in plasma/serum and acted as a biomarker in disease [29, 30]. For instance, serum OGN was an independent risk predictor for patients with chronic kidney disease [29]. This led us to speculate that OGN might serve as a serological marker for the prognosis of GC, which required to be confirmed by extra experiments. Furthermore, OGN also could adjust immune response via mediating immune cell infiltration in cancer, such as OGN expression positively associated with CD8+ T cells recruitment/infiltration by inhibited HIF-1α/ VEGF pathway in colorectal cancer [31]. Here, our results also indicated that OGN was significantly positively correlated with immune cell infiltration in GC, such as M2 macrophages. OGN might be an immune modulator in TME in GC. In a word, our study showed that OGN could be a new useful prognostic biomarker and immune regulator for GC.
CHRDL2, an antagonist of bone morphogenic proteins (BMPs), worked as an oncogene in colorectal cancer [32] and osteosarcoma [33]. CHRDL2 was firstly discovered in 2003 named as BNF-1 (breast tumor novel factor 1) and found its overexpression in breast, lung, and colon tumors detected by PCR in a small sample [34]. While the report of CHRDL2 in the tumor is very limited. Here, we first pointed out that CHRDL2 expression increased in the GC process and its high expression was associated with poor prognosis in GC patients. As a secreted protein, we found its expression associated with immune cell infiltration, especially for M2 macrophage, and immune inhibit molecular markers in GC. Therefore, CHRDL2 might be a novel target of GC therapy.
C2orf40 encodes a protein called esophageal cancer-related gene-4 (ECRG4), which is down-regulated by hypermethylation of its promoter in diverse types of tumors, including hepatocellular carcinoma [35], GC [36], breast cancer [37]. According to current reports, c2orf40 might as a potential tumor suppressor gene in tumors. However, our results exhibited that high expression of ECRG4 was remarkably associated with poor outcomes (HR = 1.95, log-rank P = 6.7e−10), and its expression increased gradually in GC advance. The opposite result may be on account of different levels of evidence or cohort studies [36, 38]. The deep reasons are yet to be further validated. It has been reported that ECRG4 interacted with TLR4 [36]. And TLR4/PI3K/Akt signaling was a vital way to promote M2 polarization of macrophages [10]. Our results exhibited C2orf40/ ECRG4 expression notably related to immune cell infiltration, including M2 macrophages. Thus, it also needs to verify whether ECRG4 is involved in the development of GC by modifying the M2 polarization of macrophages through TLR4.
THBS4 belongs to the thrombospondin protein family, which is a kind of adhesive glycoproteins that mediate cell-to-cell and cell-to-matrix interactions. THBS4 tended to be an oncogene in GC [39], colorectal cancer [40], prostate cancer [41], and hepatocellular carcinoma [42]. As an illustration, inhibited expression of THBS4 could impede the PI3K/Akt signaling and disturb the cancer stem cell (CSC)-like properties in prostate cancer [41]. And THBS4 might as a biomarker for diffuse-type gastric adenocarcinomas [43], and a potential indicator for risk assessment and prognosis prediction of GC according to its polymorphisms [44] and bioinformatics analysis [45]. In addition, THBS4 is a secreted extracellular matrix protein that has been reported to mediate angiogenesis, adhesion, migration, and proliferation responding to TGF-β signaling [46]. Our results suggested THBS4 expression was associated with TGFβ in GC. Whether THBS4 responds to TGFβ signaling pathway in GC and participates in the progression of GC remains to be verified experimentally. Our study found that the expression of THBS4 was notably increased following the advanced procession of GC and TGFβ pathways might also facilitate THBS4 secretion to promoting GC. THBS4 also positively correlated with others immunoinhibitors and majority immunostimulators. The important role of THBS4 in GC still needs more evidence to prove.
CHRDL1 is a paralog of CHRDL2, which has only been reported in two papers in GC. One paper reported that its expression associated with CLIP4 DNA methylation and CHRDL1 might be a prognostic signature gene [47]. Another reported low expression of CHRDL1, as an antagonist of bone morphogenetic protein 4 (BMP4), might promote GC cell proliferation and migration by BMP receptor II [48]. However, our results revealed that high expression of CHRDL1 with poor OS in GC by detecting in two datasets, and CHRDL1 expression was associated with tumor stages, immune cell infiltration, immunoinhibitors, and immunostimulators.
Many papers showed ANGPTL1 acted as a tumor suppressor by inhibiting angiogenesis, cancer metastasis, cancer stemness, and repressing sorafenib resistance in treatment [49, 50]. While its role in GC still not clear [51]. Our results revealed that ANGPTL1 might be a potential target in GC, according to its high expression related to poor OS and gradually increased with tumor stages as well as the feasible relationship with immune cells and biomarkers in TME.
4 Conclusions
Here, we searched a series of databases to find out the most relevant molecular that their expression closely related to the survival of GC. And functional enrichment analysis of survival-related CMS genes was performed. These genes are involved in many cancers, including GC, and cancer processes, such as angiogenesis. Then, we picked 6 genes that could be secreted in the TME and enhanced in immune cells. Further survival analysis demonstrated that high expression of OGN, CHRDL2, C2orf40, THBS4, CHRDL1, and ANGPTL1, respectively, was significantly associated with poor OS in GC patients. OGN and CHRDL2 were the firstly reported in GC. Moreover, the expression of these 6 genes prominently increased with the tumor pathological stage. In addition, we investigated the pathways of those hub genes by enrichment analysis. Ultimately, immune infiltration analysis displayed that those hub genes expression positively with CD8+ T Cell, M2 macrophage, most immunoinhibitors, and majority immunostimulators. These processes are closely relative to tumor growth and metastasis. Therefore, high expression of 6 hub genes alone or synergistically resulted in a poor prognosis in GC patients. More experiments are still needed to verify these findings.
5 Materials and methods
5.1 COMSUC
COMSUC [12] (http://comsuc.bioinforai.tech/home) is used to identify Consensus Molecular Subtypes (CMS) by integrating multiple clustering results based on multiple platforms, multiple omics data, and multiple methods. In this study, we integrated clustering results of GC data from TCGA into two groups by three algorithms based on K-means, hierarchical clustering (Hclust), non-negative matrix factorization (NMF).
5.2 Metascape
Metascape [13] (http://metascape.org/gp/index.html#/main/step1) is a web server designed to provide an extensive gene list annotation and analysis resource for users. Enrichment analysis was the essential part of Metascape. Here, we analyzed the prominent changed genes of CMS1 for enrichment analysis by custom analysis.
5.3 Kaplan–Meier plotter
Kaplan–Meier plotter [18] (http://kmplot.com/analysis/) is a platform utilized to discover and validate the survival biomarkers of four cancers, including gastric cancer. To analyze the prognostic value of OGN, CHRDL2, C2orf40, THBS4, CHRDL1, and ANGPTL1, the cohorts were divided into high- and low- groups through their expression. In this study, the overall survival of 6 hub genes was compared in GC. And the hazard ratios (HRs, with 95% confidence intervals) and log-rank P-values (< 0.05 as significant difference) were counted.
5.4 GEPIA
Gene Expression Profiling Interactive Analysis [22] (GEPIA, http://gepia.cancer-pku.cn/) uses a standard processing approach to examine genes expression by RNA sequencing data for 8587 normal samples and 9736 tumors from GTEx and TCGA projects. Here, GEPIA was used to investigate the six hub genes expression correlation for GC tumor stages from TCGA data. In addition, we valued the correlation between the expression of CSF1R/CXCL12 and six hub genes base on GEPIA.
5.5 TIMER
TIMER [23] (https://cistrome.shinyapps.io/timer/) is a web tool used to investigate immune cell infiltration in diverse cancers, including GC. It provides data to evaluate the associations between expression levels of selected genes and infiltrating immune cells, such as B cells, CD4+ T cells, CD8+ T cells, neutrophils, macrophages, and dendritic cells. In our study, we estimated the correlation of expression of OGN, CHRDL2, C2orf40, THBS4, CHRDL1, and ANGPTL1 with immune cell infiltration.
5.6 TISIDB
TISIDB [24] (http://cis.hku.hk/TISIDB/index.php) is an online web for assessing tumor-related gene and immune system interaction. In our study, we used TISIDB to find out the correlation between the expression of six hub genes and the abundance of immunomodulators. And distribution of six hub genes expression across immune and molecular subtypes in GC. The correlations between six hub genes and the immune system were measured by Spearman’s test.
Data availability
All data generated or analyzed during this study are included in this article.
Code availability
Not applicable.
Change history
20 December 2021
A Correction to this paper has been published: https://doi.org/10.1007/s12672-021-00458-x
Abbreviations
- GC:
-
Gastric cancer
- ICB:
-
Immune checkpoint blockade
- TME:
-
Tumor microenvironment
- OS:
-
Overall survival
- CMS:
-
Consensus molecular subtypes
- TFs:
-
Transcription factors
- SLRP:
-
Small leucine-rich proteoglycan
- BMPs:
-
Bone morphogenic proteins
- BNF-1:
-
Breast tumor novel factor 1
- ECRG4:
-
Esophageal cancer-related gene-4
- CSC:
-
Cancer stem cell
References
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021. https://doi.org/10.3322/caac.21660.
Allemani C, et al. Global surveillance of trends in cancer survival 2000–14 (CONCORD-3): analysis of individual records for 37 513 025 patients diagnosed with one of 18 cancers from 322 population-based registries in 71 countries. Lancet. 2018;391(10125):1023–75. https://doi.org/10.1016/S0140-6736(17)33326-3.
Shitara K, et al. Pembrolizumab versus paclitaxel for previously treated, advanced gastric or gastro-oesophageal junction cancer (KEYNOTE-061): a randomised, open-label, controlled, phase 3 trial. Lancet. 2018;392(10142):123–33. https://doi.org/10.1016/S0140-6736(18)31257-1.
Reiser J, Banerjee A. Effector, memory, and dysfunctional CD8(+) T cell fates in the antitumor immune response. J Immunol Res. 2016;2016:8941260. https://doi.org/10.1155/2016/8941260.
Wang Y, Zhu C, Song W, Li J, Zhao G, Cao H. PD-L1 expression and CD8(+) T cell infiltration predict a favorable prognosis in advanced gastric cancer. J Immunol Res. 2018;2018:4180517. https://doi.org/10.1155/2018/4180517.
Shen Y, Teng Y, Lv Y, Zhao Y, Qiu Y, Chen W, Wang L, Wang Y, Mao F, Cheng P, Ma D, Zhuang Y, Zou Q, Peng L. PD-1 does not mark tumor-infiltrating CD8+ T cell dysfunction in human gastric cancer. J Immunother Cancer. 2020. https://doi.org/10.1136/jitc-2019-000422.
Hashimoto M, Kamphorst AO, Im SJ, Kissick HT, Pillai RN, Ramalingam SS, Araki K, Ahmed R. CD8 T cell exhaustion in chronic infection and cancer: opportunities for interventions. Annu Rev Med. 2018;69:301–18. https://doi.org/10.1146/annurev-med-012017-043208.
Wang J, Li R, Cao Y, Gu Y, Fang H, Fei Y, Lv K, He X, Lin C, Liu H, Zhang H, Li H, He H, Xu J, Haung H. Intratumoral CXCR5(+)CD8(+)T associates with favorable clinical outcomes and immunogenic contexture in gastric cancer. Nat Commun. 2021;12(1):3080. https://doi.org/10.1038/s41467-021-23356-w.
Zhou Z, Xia G, Xiang Z, Liu M, Wei Z, Yan J, Chen W, Zhu J, Awasthi N, Sun X, Fung KM, He Y, Li M, Zhang CA. C-X-C chemokine receptor type 2-dominated cross-talk between tumor cells and macrophages drives gastric cancer metastasis. Clin Cancer Res. 2019;25(11):3317–28. https://doi.org/10.1158/1078-0432.CCR-18-3567.
Li Q, Wu W, Gong D, Shang R, Wang J, Yu H. Propionibacterium acnes overabundance in gastric cancer promote M2 polarization of macrophages via a TLR4/PI3K/Akt signaling. Gastric Cancer. 2021. https://doi.org/10.1007/s10120-021-01202-8.
Kim KJ, Wen XY, Yang HK, Kim WH, Kang GH. Prognostic implication of M2 macrophages are determined by the proportional balance of tumor associated macrophages and tumor infiltrating lymphocytes in microsatellite-unstable gastric carcinoma. PLoS ONE. 2015;10(12): e0144192. https://doi.org/10.1371/journal.pone.0144192.
He S, Song X, Yang X, Yu J, Wen Y, Wu L, Yan B, Feng J, Bo X. COMSUC: a web server for the identification of consensus molecular subtypes of cancer based on multiple methods and multi-omics data. PLoS Comput Biol. 2021;17(3): e1008769. https://doi.org/10.1371/journal.pcbi.1008769.
Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, Benner C, Chanda SK. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10(1):1523. https://doi.org/10.1038/s41467-019-09234-6.
Wu H, Liu X, Gong P, Song W, Zhou M, Li Y, Zhao Z, Fan H. Elevated TFAP4 regulates lncRNA TRERNA1 to promote cell migration and invasion in gastric cancer. Oncol Rep. 2018;40(2):923–31. https://doi.org/10.3892/or.2018.6466.
Wadhwa R, Song S, Lee JS, Yao Y, Wei Q, Ajani JA. Gastric cancer-molecular and clinical dimensions. Nat Rev Clin Oncol. 2013;10(11):643–55. https://doi.org/10.1038/nrclinonc.2013.170.
Liu H, Du F, Sun L, Wu Q, Wu J, Tong M, Wang X, Wang Q, Cao T, Gao X, Cao J, Wu N, Nie Y, Fan D, Lu Y, Zhao X. GATA6 suppresses migration and metastasis by regulating the miR-520b/CREB1 axis in gastric cancer. Cell Death Dis. 2019;10(2):35. https://doi.org/10.1038/s41419-018-1270-x.
Ashrafizadeh M, Najafi M, Ang HL, Moghadam ER, Mahabady MK, Zabolian A, Jafaripour L, Bejandi AK, Hushmandi K, Saleki H, Zarrabi A, Kumar AP. PTEN, a barrier for proliferation and metastasis of gastric cancer cells: from molecular pathways to targeting and regulation. Biomedicines. 2020. https://doi.org/10.3390/biomedicines8080264.
Nagy A, Munkacsy G, Gyorffy B. Pancancer survival analysis of cancer hallmark genes. Sci Rep. 2021;11(1):6047. https://doi.org/10.1038/s41598-021-84787-5.
Xia Y, Wang L, Xu Z, Kong R, Wang F, Yin K, Xu J, Li B, He Z, Wang L, Xu H, Zhang D, Yang L, Wu JY, Xu Z. Reduced USP33 expression in gastric cancer decreases inhibitory effects of Slit2-Robo1 signalling on cell migration and EMT. Cell Prolif. 2019;52(3): e12606. https://doi.org/10.1111/cpr.12606.
Shi R, Yang Z, Liu W, Liu B, Xu Z, Zhang Z. Knockdown of Slit2 promotes growth and motility in gastric cancer cells via activation of AKT/beta-catenin. Oncol Rep. 2014;31(2):812–8. https://doi.org/10.3892/or.2013.2887.
Aanaya J. OncoLnc: linking TCGA survival data to mRNAs, miRNAs, and lncRNAs. PeerJ Comput Sci. 2016;2:e67. https://doi.org/10.7717/peerj-cs.67.
Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–102. https://doi.org/10.1093/nar/gkx247.
Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, Li B, Liu XS. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77(21):e108–10. https://doi.org/10.1158/0008-5472.CAN-17-0307.
Ru B, Wong CN, Tong Y, Zhong JY, Zhong SSW, Wu WC, Chu KC, Wong CY, Lau CY, Chen I, Chan NW, Zhang J. TISIDB: an integrated repository portal for tumor-immune system interactions. Bioinformatics. 2019;35(20):4200–2. https://doi.org/10.1093/bioinformatics/btz210.
Cancer Genome Atlas Research N. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513(7517):202–9. https://doi.org/10.1038/nature13480.
Liu Y, et al. Comparative molecular analysis of gastrointestinal adenocarcinomas. Cancer Cell. 2018;33(4):721-35 e8. https://doi.org/10.1016/j.ccell.2018.03.010.
Mei Y, Du Z, Hu C, Greenwald NF, Abedalthagafi M, Agar NYR, Dunn GP, Bi WL, Santagata S, Dunn IF. Osteoglycin promotes meningioma development through downregulation of NF2 and activation of mTOR signaling. Cell Commun Signal. 2017;15(1):34. https://doi.org/10.1186/s12964-017-0189-7.
Hu X, Li YQ, Li QG, Ma YL, Peng JJ, Cai SJ. Osteoglycin (OGN) reverses epithelial to mesenchymal transition and invasiveness in colorectal cancer via EGFR/Akt pathway. J Exp Clin Cancer Res. 2018;37(1):41. https://doi.org/10.1186/s13046-018-0718-2.
Baek SH, Cha RH, Kang SW, Park CW, Cha DR, Kim SG, Yoon SA, Kim S, Han SY, Park JH, Chang JH, Lim CS, Kim YS, Na KY. Higher serum levels of osteoglycin are associated with all-cause mortality and cardiovascular and cerebrovascular events in patients with advanced chronic kidney disease. Tohoku J Exp Med. 2017;242(4):281–90. https://doi.org/10.1620/tjem.242.281.
Seki T, Saita E, Kishimoto Y, Ibe S, Miyazaki Y, Miura K, Ohmori R, Ikegami Y, Kondo K, Momiyama Y. Low levels of plasma osteoglycin in patients with complex coronary lesions. J Atheroscler Thromb. 2018;25(11):1149–55. https://doi.org/10.5551/jat.43059.
Hu X, Li YQ, Li QG, Ma YL, Peng JJ, Cai SJ. Osteoglycin-induced VEGF inhibition enhances T lymphocytes infiltrating in colorectal cancer. EBioMedicine. 2018;34:35–45. https://doi.org/10.1016/j.ebiom.2018.07.021.
Sun J, Liu X, Gao H, Zhang L, Ji Q, Wang Z, Zhou L, Wang Y, Sui H, Fan Z, Li Q. Overexpression of colorectal cancer oncogene CHRDL2 predicts a poor prognosis. Oncotarget. 2017;8(7):11489–506. https://doi.org/10.18632/oncotarget.14039.
Chen H, Pan R, Li H, Zhang W, Ren C, Lu Q, Chen H, Zhang X, Nie Y. CHRDL2 promotes osteosarcoma cell proliferation and metastasis through the BMP-9/PI3K/AKT pathway. Cell Biol Int. 2021;45(3):623–32. https://doi.org/10.1002/cbin.11507.
Wu I, Moses MA. BNF-1, a novel gene encoding a putative extracellular matrix protein, is overexpressed in tumor tissues. Gene. 2003;311:105–10. https://doi.org/10.1016/s0378-1119(03)00563-8.
Wu Y, Xiang Q, Lv X, Xiang X, Feng Z, Tian S, Tang J, Xiang T, Gong J. C2orf40 inhibits hepatocellular carcinoma through interaction with UBR5. J Gastroenterol Hepatol. 2021. https://doi.org/10.1111/jgh.15441.
Deng P, Chang XJ, Gao ZM, Xu XY, Sun AQ, Li K, Dai Q. Downregulation and DNA methylation of ECRG4 in gastric cancer. Onco Targets Ther. 2018;11:4019–28. https://doi.org/10.2147/OTT.S161200.
Tang GY, Tang GJ, Yin L, Chao C, Zhou R, Ren GP, Chen JY, Zhang W. ECRG4 acts as a tumor suppressor gene frequently hypermethylated in human breast cancer. 2019. Biosci Rep. https://doi.org/10.1042/BSR20190087.
Chen L, Tang H, Liu G, Xiao S, Liang D, Ma J, Yang Y, Luo H, Zhu Y, Xie F, Cheng X, Chi J, Wu X. MicroRNA-196b promotes gastric cancer progression by targeting ECRG4. Anticancer Drugs. 2021;32(2):127–37. https://doi.org/10.1097/CAD.0000000000000998.
Chen X, Huang Y, Wang Y, Wu Q, Hong S, Huang Z. THBS4 predicts poor outcomes and promotes proliferation and metastasis in gastric cancer. J Physiol Biochem. 2019;75(1):117–23. https://doi.org/10.1007/s13105-019-00665-9.
Kim MS, Choi HS, Wu M, Myung J, Kim EJ, Kim YS, Ro S, Ha SE, Bartlett A, Wei L, Ryu HS, Choi SC, Park WC, Kim KY, Lee MY. Potential role of PDGFRbeta-associated THBS4 in colorectal cancer development. Cancers (Basel). 2020. https://doi.org/10.3390/cancers12092533.
Hou Y, Li H, Huo W. THBS4 silencing regulates the cancer stem cell-like properties in prostate cancer via blocking the PI3K/Akt pathway. Prostate. 2020;80(10):753–63. https://doi.org/10.1002/pros.23989.
Guo D, Zhang D, Ren M, Lu G, Zhang X, He S, Li Y. THBS4 promotes HCC progression by regulating ITGB1 via FAK/PI3K/AKT pathway. FASEB J. 2020;34(8):10668–81. https://doi.org/10.1096/fj.202000043R.
Forster S, Gretschel S, Jons T, Yashiro M, Kemmner W. THBS4, a novel stromal molecule of diffuse-type gastric adenocarcinomas, identified by transcriptome-wide expression profiling. Mod Pathol. 2011;24(10):1390–403. https://doi.org/10.1038/modpathol.2011.99.
Lin X, Hu D, Chen G, Shi Y, Zhang H, Wang X, Guo X, Lu L, Black D, Zheng XW, Luo X. Associations of THBS2 and THBS4 polymorphisms to gastric cancer in a Southeast Chinese population. Cancer Genet. 2016;209(5):215–22. https://doi.org/10.1016/j.cancergen.2016.04.003.
Lu Y, Kong X, Zhong W, Hu M, Li C. Diagnostic, therapeutic, and prognostic value of the thrombospondin family in gastric cancer. Front Mol Biosci. 2021;8: 647095. https://doi.org/10.3389/fmolb.2021.647095.
Muppala S, Xiao R, Krukovets I, Verbovetsky D, Yendamuri R, Habib N, Raman P, Plow E, Stenina-Adognravi O. Thrombospondin-4 mediates TGF-beta-induced angiogenesis. Oncogene. 2017;36(36):5189–98. https://doi.org/10.1038/onc.2017.140.
Hu C, Zhou Y, Liu C, Kang Y. A novel scoring system for gastric cancer risk assessment based on the expression of three CLIP4 DNA methylation-associated genes. Int J Oncol. 2018;53(2):633–43. https://doi.org/10.3892/ijo.2018.4433.
Pei YF, Zhang YJ, Lei Y, Wu WD, Ma TH, Liu XQ. Hypermethylation of the CHRDL1 promoter induces proliferation and metastasis by activating Akt and Erk in gastric cancer. Oncotarget. 2017;8(14):23155–66. https://doi.org/10.18632/oncotarget.15513.
Yan Q, Jiang L, Liu M, Yu D, Zhang Y, Li Y, Fang S, Li Y, Zhu YH, Yuan YF, Guan XY. ANGPTL1 interacts with integrin alpha1beta1 to suppress HCC angiogenesis and metastasis by inhibiting JAK2/STAT3 signaling. Cancer Res. 2017;77(21):5831–45. https://doi.org/10.1158/0008-5472.CAN-17-0579.
Chen HA, Kuo TC, Tseng CF, Ma JT, Yang ST, Yen CJ, Yang CY, Sung SY, Su JL. Angiopoietin-like protein 1 antagonizes MET receptor activity to repress sorafenib resistance and cancer stemness in hepatocellular carcinoma. Hepatology. 2016;64(5):1637–51. https://doi.org/10.1002/hep.28773.
Tang C, Chen E, Peng K, Wang H, Cheng X, Wang Y, Yu S, Yu Y, Cui Y, Liu T. Mining the role of angiopoietin-like protein family in gastric cancer and seeking potential therapeutic targets by integrative bioinformatics analysis. Cancer Med. 2020;9(13):4850–63. https://doi.org/10.1002/cam4.3100.
Acknowledgements
This study is supported partially by the National Natural Science Foundation of China (grant numbers 82001159), the Science and Technology program of Jiangxi Provincial health commission (grant numbers 202130212), The Science and technology research project of the Education Department of Jiangxi Province (grant numbers GJJ200229), Technology Program of Jiangxi Administration of Traditional Chinese Medicine (grant numbers 2020A0247).
Author information
Authors and Affiliations
Contributions
MM, BY, and XY performed the experimental design and data analysis. MM wrote the manuscript. MM, XY, BY, and WF revised the manuscript. JX supervised the study. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
The study was approved by the Ethics Committee of The First Affiliated Hospital of Nanchang University.
Consent for publication
Not applicable.
Competing interests
The authors have no conflicts of interest to disclose.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article has been updated to provide a different reference 16 as the original source was retracted.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yu, X., Yu, B., Fang, W. et al. Identification hub genes of consensus molecular subtype correlation with immune infiltration and predict prognosis in gastric cancer. Discov Onc 12, 41 (2021). https://doi.org/10.1007/s12672-021-00434-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12672-021-00434-5