Abstract
Gene expression in blood was correlated with mercury levels in blood of 2- to 5-year-old boys with autism (AU) compared to age-matched typically developing (TD) control boys. This was done to address the possibility that the two groups might metabolize toxicants, such as mercury, differently. RNA was isolated from blood and gene expression assessed on whole genome Affymetrix Human U133 expression microarrays. Mercury levels were measured using an inductively coupled plasma mass spectrometer. Analysis of covariance (ANCOVA) was performed and partial correlations between gene expression and mercury levels were calculated, after correcting for age and batch effects. To reduce false positives, only genes shared by the ANCOVA models were analyzed. Of the 26 genes that correlated with mercury levels in both AU and TD boys, 11 were significantly different between the groups (P(Diagnosis*Mercury) ≤ 0.05). The expression of a large number of genes (n = 316) correlated with mercury levels in TD but not in AU boys (P ≤ 0.05), the most represented biological functions being cell death and cell morphology. Expression of 189 genes correlated with mercury levels in AU but not in TD boys (P ≤ 0.05), the most represented biological functions being cell morphology, amino acid metabolism, and antigen presentation. These data and those in our companion study on correlation of gene expression and lead levels show that AU and TD children display different correlations between transcript levels and low levels of mercury and lead. These findings might suggest different genetic transcriptional programs associated with mercury in AU compared to TD children.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Autism (AU) is a neurodevelopmental disorder characterized by impaired social interactions and impaired verbal and nonverbal communication as well as restricted, stereotyped patterns of behavior and interests. The etiology(ies) of autism remains elusive. The interactions of multiple genetic, environmental, and epigenetic factors are postulated to be involved (Ashwood et al. 2006; Hertz-Picciotto et al. 2006; London 2000). Exposures to heavy metals have received much attention because of their developmental neurotoxicity (WHO 1990, 1991; Tchounwou et al. 2003; Clarkson and Magos 2006). Mercury is a toxic and hazardous heavy metal that is ubiquitous in the environment, being found in fish, thermometers, dental amalgams, and in the atmosphere and affects multiple organ systems (WHO 1990, 1991; Tchounwou et al. 2003; Clarkson and Magos 2006; Counter and Buchanan 2004; Zahir et al. 2005). In animals, mercury binds to the endogenous tripeptide glutathione (GSH), and the resulting mercury–glutathione complex is metabolized by the ectoproteins gamma-glutamyl transpeptidase (GGT) and dipeptidase (Rice 2008; Newland et al. 2008; Guzzi and La Porta 2008). Mercury is then eliminated as GSH conjugates, GSH being produced by glutamyl-cysteine ligase (GCL) and conjugation by glutathione S-transferases (GST). The high affinity of mercury for sulfhydryl, and to a lesser degree hydroxyl, carboxyl, and phosphoryl groups of structural and functional proteins, is believed to be the biochemical basis for its harmful effects on biological systems (Clarkson and Magos 2006).
The element mercury exists in three species: elemental mercury (Hgo), inorganic mercury compounds (primarily mercuric chloride), and organic mercury (primarily methylmercury). Exposure to each mercury species can cause general as well as specific toxic effects in children and adults (Clarkson and Magos 2006; Counter and Buchanan 2004). For example, methylmercury derived mostly from fish consumption passes through the blood–brain barrier as an l-cysteine complex via the l-amino acid transporter (Clarkson and Magos 2006; Mokrzan et al. 1995). The mechanisms by which inorganic mercury damages human brain, however, are poorly understood. The different mercury species have different absorption, distribution, metabolism, excretion, and toxicity (Guzzi and La Porta 2008; Ercal et al. 2001; Quig 1998). In addition, the effect of a toxicant can be influenced by another substance, so-called “mixture toxicity” (Rice 2008; Newland et al. 2008; McCarty and Borgert 2006a, b).
The development of whole genome microarrays has made it possible to characterize how the entire transcriptome of cells or organisms responds to changes in external or internal environments. Measurements of gene expression (RNA) levels provide a “transcriptional signature” of an individual’s genetic, physiological, and environmental status. The relationship between levels of xenobiotics and transcriptional signatures measured in blood can be used to provide mechanistic information about toxic exposures (Yoon et al. 2008).
The objective of the current study was to analyze the entire transcriptome in similar aged typically developing (TD) and AU boys to identify those genes whose expression level in blood correlated with relatively low total mercury (Hg) levels in blood. Moreover, the correlations of RNA levels with Hg levels might differ in AU compared to TD boys. Lastly, we postulated that differences in RNA expression signatures for AU compared to TD boys would occur not only for Hg but would also be different for other xenobiotics including lead (Tian et al. 2009, accompanying paper). This study begins to address these hypotheses.
Materials and Methods
Subjects
Subjects were participants in the CHARGE study (Hertz-Picciotto et al. 2006). These children were assessed at the UC Davis Medical Investigations of Neurodevelopmental Disorders (M.I.N.D.) Institute. Each child was evaluated using standardized clinical assessments. AU cases were assessed using the Autism Diagnostic Interview–Revised (ADI-R) (Le Couteur et al. 2003; Lord et al. 1994, 1997) and the Autism Diagnostic Observation Schedules (ADOS) (Lord et al. 2000, 2003). For this study of gene expression versus Hg levels, we analyzed boys with AU (n = 33, mean age 45.3 with a range of 31–60 months) and age-matched TD control boys (n = 51, mean age 43.3 with a range of 28–57 months). Only subjects for whom the Affymetrix expression data was generated in 2007–2008 were analyzed because Affymetrix arrays processed prior to those times were done using a different protocol which resulted in significant technical variation. Only boys were analyzed in our study because there were few girls who had both Affymetrix expression data and blood Hg data. The subjects used in our companion study on correlation between gene expression and blood lead levels are different from the ones used in this study. Parents provided informed consent for all subjects (Tian et al. 2009). The study was approved by the Institutional Review Board at University of California Davis Medical Center and was conducted in accordance with the Declaration of Helsinki.
Hg Analysis
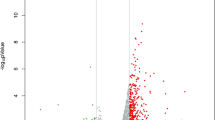
Total blood Hg was measured on an Agilent 7500i Inductively Coupled Plasma Mass Spectrometer (ICP-MS) (Agilent, Palo Alto, CA) in the UCD Department of Civil and Environmental Engineering. Detailed methods are included in our previous publication (Hertz-Picciotto et al. 2009). Blood Hg concentrations were log2-transformed due to the skewed distribution over a wide range of values (Fig. 2). The normality of the distributions was assessed by performing the Kolmogorov–Smirnov test in Partek Genomics Suite 6.4. The detection limit for Hg was 0.02 μg/l (Hertz-Picciotto et al. 2009), with the (slightly) lower detection limit of 0.01 μg/l (limit of barely detected). Log2-transformation of the Hg levels was performed to produce a more linear distribution of the values and to match the log2-transformation of gene expression (see below). For log2-transformation of the Hg data, Hg levels below detection levels were assigned a value of 0.009 μg/l. The value of 0.009 was selected to be slightly lower than the lower limit of Hg detection of 0.01 μg/l in order to not create “outlying” values, which can artificially influence the correlation coefficient.
RNA Isolation and Affymetrix Array Hybridizations
Whole blood was collected and total RNA was isolated using PAXgene tubes and kits (Qiagen, Valencia, CA). RNA concentration and purity were checked using a Nanodrop ND-1000 spectrophotometer, and integrity was checked using the Agilent 2100 Bioanalyzer (260/280>2.0; 28S/18S>1.5; RIN>8).
Human whole genome U133 Plus 2.0 GeneChip microarrays (Affymetrix; Santa Clara, CA), surveying over 54,000 probe sets, were used. The 54,000 probe sets represent over 38,500 potential human genes (Affymetrix Manual). The standard Affymetrix protocol was followed for the sample labeling, hybridization, and image scanning.
Statistical Analysis
Analysis of Affymetrix Expression Array Data
Raw gene expression values from Affymetrix.cel files were imported into Partek Genomics Suit, version 6.4, release 6.09.0422 (Partek Inc., St. Louis, MI). Probe summarization and probe set normalization were performed using the GC-RMA algorithm (Wu and Irizarry 2004), including GC-RMA background correction, Quantile Normalization, log2-transformation, and Median Polish probe set summarization.
Analysis of Covariance
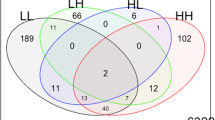
Analysis of covariances (ANCOVAs) were performed in Partek Genomics Suite, version 6.4, release 6.09.0422 (Partek Inc., St. Louis, MI). We estimated the partial correlations between log2-transformed gene expression and log2-transformed Hg levels while removing the effects of age and microarray technical variation (batch) (Eisenhart 1947). Partial correlation coefficients describe the linear relation between two variables (in our case, gene expression and Hg level) while controlling for the effects of one or more additional variables (Morrison 1976). Correlations are a measure of linear association. A positive partial correlation between Hg level and gene expression indicates that gene expression is increasing when Hg levels are increasing. Similarly, a negative partial correlation indicates that gene expression is decreasing when Hg levels are increasing. We performed the ANCOVA on several models (see below). We overlapped these gene lists (Fig. 1) to reduce the large false discovery rates due to multiple comparisons between thousands of genes. The following ANCOVA models were used.
Analysis design—Venn diagram of the three ANCOVA models used to generate the genes significantly correlating with mercury (Hg) levels. GE gene expression, TD typically developing control children from the general population, AU children with autism, Batch a group of microarrays that were processed together. Gene List A genes whose expression correlates with Log2Hg levels in each of the TD and AU models separately (P(Log2Hg) ≤ 0.05), with no significant differences in the correlations between the two groups in the combined AU + TD model (P(Diagnosis*Log2Hg) > 0.05). Gene List B genes whose expression correlates with Log2Hg levels in each of the TD and AU models separately (P(Log2Hg) ≤ 0.05), and had significant differences in the correlations between the two groups in the combined AU + TD model (P(Diagnosis*Log2Hg) ≤ 0.05). Gene List C genes whose expression correlates with Log2Hg levels in the TD model (P(Log2Hg) ≤ 0.05) but not in the AU model (P(Log2Hg) > 0.05), and which had significant differences in the correlations between the two groups in the combined AU + TD model (P(Diagnosis*Log2Hg) ≤ 0.05). Gene List D represents genes whose expression correlates with Log2Hg levels in the AU model (P(Log2Hg) ≤ 0.05) but not in the TD model (P(Log2Hg) > 0.05), and which had significant differences in the correlations between the two groups in the combined AU + TD model (P(Diagnosis*Log2Hg) ≤ 0.05)
Model 1: TD Boys Only
Log2(Gene Expression) is a function of the following factors: Log2Hg (continuous), Age (continuous), and Batch (categorical). Genes with a P(Log2(Hg)) ≤ 0.05 in this model would be associated with the left circle in Fig. 1.
Model 2: AU Boys Only
Log2(Gene Expression) is a function of the following factors: Log2Hg (continuous), Age (continuous), and Batch (categorical). Genes with a P(Log2(Hg)) ≤ 0.05 in this model would be associated with the right circle in Fig. 1.
Model 3: AU and TD Boys Combined
Log2(Gene Expression) is a function of the following factors: Log2Hg (continuous), Age (continuous), Batch (categorical), Diagnosis (binary categorical), and interaction Diagnosis*Log2Hg.
Genes with a P(Diagnosis*Log2Hg) ≤ 0.05 would be associated with the top circle in Fig. 1.
Batch in all three models is a random effect. We focused on gene lists that came from the combined analyses to minimize the false discovery rate (Fig. 1, intersections labeled A–D).
-
Gene list A represents genes whose expression correlates with Log2Hg levels in each of the TD and AU models separately (P(Log2Hg) ≤ 0.05), with no significant differences in the correlations between the two groups in the combined AU + TD model (P(Diagnosis*Log2Hg) > 0.05).
-
Gene list B represents genes whose expression correlates with Log2Hg levels in each of the TD and AU models separately (P(Log2Hg) ≤ 0.05), and had significant differences in the correlations between the two groups in the combined AU + TD model (P(Diagnosis*Log2Hg) ≤ 0.05).
-
Gene list C represents genes whose expression correlates with Log2Hg levels in the TD model (P(Log2Hg) ≤ 0.05) but not in the AU model (P(Log2Hg) > 0.05), and which had significant differences in the correlations between the two groups in the combined AU + TD model (P(Diagnosis*Log2Hg) ≤ 0.05).
-
Gene list D represents genes whose expression correlates with Log2Hg levels in the AU model (P(Log2Hg) ≤ 0.05) but not in the TD model (P(Log2Hg) > 0.05), and which had significant differences in the correlations between the two groups in the combined AU + TD model (P(Diagnosis*Log2Hg) ≤ 0.05).
Assessment of Possible Biological Significance of Identified Genes
Gene ontology annotation was obtained from the Affymetrix NetAffix database (www.affymetrix.com). Pathway analysis was performed using Ingenuity Pathways Analysis (IPA) (Ingenuity Systems®, www.ingenuity.com). A dataset containing gene identifiers (called focus molecules) from each of the lists (A, B, C, or D) was uploaded into the program. Each identifier was mapped to its corresponding gene object and overlaid onto a global molecular network developed from information contained in the Ingenuity knowledge base. The Bio Functional Analysis identified the biological functions and/or diseases that were most significant to the dataset. Canonical Pathway Analysis identified the pathways from the IPA library of canonical pathways that were most significant to the dataset. The significance of the association between the dataset and canonical pathways was assessed by calculating the ratio of the number of genes from the dataset that map to the pathway divided by the total number of molecules that exist in the canonical pathway. A Right-tailed Fischer’s Exact Test was then used to calculate a P-value determining the probability that each biological function and/or disease assigned to that dataset is due to chance alone. A P < 0.05 was considered to be statistically significant for over-representation of the molecules in a given process. Thus, over-represented functional or pathway processes are processes which have more focus molecules than expected by chance. It should be noted that for lists A and B (Figs. 1, 3), due to the small number of genes, pathway over-representation is not possible. In this case, IPA was used to map the genes into their corresponding pathways and biofunctions. Genomatix software (http://www.genomatix.com) was utilized to identify over-represented transcription factor binding sites in each of the four gene lists (A–D, Fig. 1).
Chromosome and cytoband enrichment for each of the gene lists (lists A, B, C, and D) as well as functional annotation of the probe sets in the enriched chromosomes was performed in DAVID (http://david.abcc.ncifcrf.gov/tools.jsp). A modified Fisher Exact P-value (the threshold of EASE score) for gene enrichment analysis was used. We considered the default P ≤ 0.1 to be considered strongly enriched in the annotation categories. A default value of 2 for the threshold of the minimum gene counts belonging to an annotation term was adopted. Visual representation of the statistically enriched chromosomes was done using Partek and was presented in Supplementary figures 1–3.
Results
Hg Levels
Total blood Hg levels ranged from below the lower detection limit (1 sample with [Hg] < 0.01 μg/l) to 4.3 μg/l in the TD group, and from below the lower detection limit (4 samples with [Hg] < 0.01 μg/l) to 3.0 μg/l in the AU group (Fig. 2). The distribution of the linear Hg levels was significantly different from the normal distribution (Kolmogorov–Smirnov test: P-value = 3.73E−04, 7.58E−03, and 6.28E−05 for TD, AU, and TD + AU, respectively). However, the distribution of the log2-transformed Hg levels followed a normal distribution (Kolmogorov–Smirnov test: P = 0.90, 0.97, and 0.94 for TD, AU, and TD + AU, respectively). There was no significant difference in Hg levels between the TD and AU groups (P = 0.42 and P = 0.07, two-sample unpaired t-test, unequal variances on the linear Hg levels and the log2-transformed Hg levels, respectively). The mean (±SD) Hg level was 0.60 (±0.82) and 0.46 (±0.73) μg/l for TD and AU, respectively. The geometric mean Hg level was 0.29 and 0.16 μg/l for TD and AU, respectively. The median was 0.30 and 0.16 μg/l for TD and AU, respectively. The inter-quartile range was 0.14–0.67 and 0.07–0.54 μg/l for TD and AU, respectively. The correlative analysis of gene expression and Hg levels were performed on log2-transformed data.
Mercury levels in children with autism (AU) and in typically developing control children from the general population (TD). The X-axis shows the samples. The Y-axis on the left shows the mercury (Hg) levels in μg/l and the Y-axis on the right shows the log2Hg levels. Symbols show Hg levels in AU (black), Hg levels in TD (gray); and circles show Hg levels (μg/l) and triangles show log2 Hg values (log2Hg)
Genes Whose Expression Correlated with Hg Levels in TD and AU Boys
ANCOVA analyses, in which the TD boys and AU boys were modeled separately, revealed 1,293 and 1,276 probe sets, respectively. The expression levels for these genes significantly correlated with Hg levels (P ≤ 0.05, Fig. 3, bottom left and right circles). Similarly, an ANCOVA analysis, in which TD and AU boys were combined, revealed 1,113 genes that correlated with Hg levels, with P ≤ 0.05 for the interaction Diagnosis (TD, AU)*log2Hg (Fig. 3, top circle). No genes passed a False-Discovery Rate filter in any of the models. Therefore, to reduce the false discovery inherent to microarray studies, we focused our analyses on the genes that were identified in more than one of the analytical models (intersections in Fig. 3, denoted as lists A–D). This approach reduced the numbers of genes approximately 4–6 times.
Numbers of genes that significantly correlate (P ≤ 0.05) with Hg levels: in children with autism (AU); and in typically developing control children from the general population (TD). The analyses used to produce the gene lists for A, B, C, and D are shown in Fig. 1. Three ANCOVAs were performed for TD, AU, and TD + AU and the numbers of significant genes are provided for each comparison and area of overlap
Genes Whose Expression Levels Correlated with Hg Levels and Exhibited a Similar Trend in Both TD and AU
There were 15 probe sets whose expression correlated with Hg levels in both the AU and TD groups, and for which the correlation coefficients did not significantly differ between the two groups (Fig. 3, list A). Figure 4 shows a heat map of the partial correlation coefficients with Hg levels for each of the 15 probe sets (rows) in the TD and AU groups (columns). Most of the probe sets showed same sign correlations with Hg levels both in the TD and AU groups. Exceptions were LOC10012902 and TXNDC17 (refer to abbreviation list for full gene name), which showed different sign correlations with Hg in the TD and AU groups. The P-values for their interaction (Diagnoses*log2Hg) were 0.06 and 0.14 which are higher than the statistical cutoff of P ≤ 0.05. This is why they were not grouped into list B (most contrasting genes). The 15 probe sets (Supplementary Table 1) represent 10 annotated genes which are involved mainly in cell death (CADM1, MSH6, SSTR3, TXNDC17, P-value range 9.92E−04 to 4.40E−02), signaling (TXNDC17, SSTR3, FEZ2), the immune response (PXDN, CADM1), and response to oxidative stress (PXDN) (Table 1). Analysis of the promoter sequences of these genes using Genomatix software revealed binding site sequences for the transcription factor SP1F family (P = 0.032, minimum 86% of sequences). The target genes are MSH6, SSTR3, PXDN, and LMLN. Chromosome 2 showed significant enrichment with 4 out of the 15 probe sets (Table 2, Supplementary Fig. 1).
Genes Whose Expression Significantly Correlated with Hg Levels but have Opposite Trends in TD and AU Boys
There were 11 probe sets, whose expression correlated with Hg levels but in opposite directions for TD compared to AU (Fig. 3, list B). Figure 5 shows a heat map of the partial correlation coefficients with Hg levels for each of the 11 probe sets (rows) in the TD and AU groups (columns). The majority of the genes show positive correlations with Hg levels in the TD group but negative correlation with Hg levels in the AU group. The 11 probe sets (Supplementary Table 1) represent seven annotated genes associated with cellular assembly and organization (NTRK3, FLNB, NCAPD3, P-value range 3.31E−04 to 2.91E−02), cellular compromise (NTRK3, FLNB, P-value range 6.62E−04 to 1.32E−03), small molecule biochemistry (KRT1, NTRK3, P-value range 9.92E−04 to 3.07E−02), vitamin and mineral metabolism (KRT1, P-value = 9.92E−04), and cell death (NTRK3, P-value = 1.35E−02) (Table 1). Analysis of the promoter sequences of these genes using Genomatix software revealed binding site sequences for the transcription factor MYBL family (P = 0.0069, minimum 86% of sequences) and the AP1R family (P = 0.0289, minimum of 86% of sequences). The target genes are FLNB and NTRK3. No significant chromosome nor cytoband enrichment (at P ≤ 0.1) was observed.
Genes correlating with Hg levels both in AU and TD, except that the correlation coefficients are significantly different from each other (List B). Heat map of the partial correlation coefficients (numbers in each box) of the gene expression and Hg levels in TD and AU. Genes are indicated in rows, and diagnoses (TD or AU) are indicated in the columns
Genes Whose Expression Correlated with Hg Levels in TD but not in AU Boys
There were 316 probe sets (Supplementary Table 1) whose expression correlated with Hg levels in the TD group but not in the AU group (Fig. 3, list C). Of these, 96 were positively correlated with Hg levels and 220 were negatively correlated (Supplementary Table 1). Figure 6 shows a heat map of the ten highest positive and ten lowest negative partial correlation coefficients with Hg levels in the TD group compared to their partial correlation coefficients in the AU group. The most represented molecular and cellular functions were cell death (37 molecules, P-value range 6.09E−04 to 4.98E−02), cell morphology (27 molecules, P-value range 9.68E−04 to 4.55E−02), gene expression (39 molecules, P-value range 2.67E−03 to 3.43E−02), cellular assembly and organization (29 molecules, P-value range 5.34E−03 to 4.98E−02), and cell cycle (17 molecules, P-value range 5.39E−03 to 4.55E−02) (Table 1). Analysis of the promoter sequences of these genes using Genomatix software revealed binding site sequences for the transcription factor ETSF family (P = 0.072, minimum 86% of sequences). Chromosomes 19, 7, and 13 showed significant enrichment with 30, 20, and 10 out of the 316 probe sets, respectively (Table 2, Supplementary Fig. 2). Functional annotation of the genes on chromosome 19 showed that they are involved in nucleic acid binding (43.3%, P = 0.01) and protein binding (60%, P = 0.09), whereas the genes on chromosome 7 have protein serine/threonine kinase activity (13%, P = 0.05). Functional annotation of the genes on chromosome 13 did not show significant over-representation of molecular functions (at P ≤ 0.1). Cytoband enrichment was noted for 22q12.1 (3/316, P = 0.03), 18q12-q21 (2/316, P = 0.04), 12q24.33 (3/316, P = 0.06), and 19q13.4 (4/316, P = 0.08).
Genes with the ten highest positive and the ten lowest negative correlations of gene expression with Hg levels for the TD subjects (List C). Heat map of the partial correlation coefficients (numbers in the boxes) of the gene expression and Hg levels in TD and AU. Genes are on the Y-axis, and diagnoses are on the X-axis
Genes Whose Expression Correlated with Hg Levels in AU but not in TD Boys
There were 189 probe sets (Supplementary Table 1) whose expression correlated with Hg levels in the AU group but not in the TD group (Fig. 3, list D). Of these, 62 were positively correlated with Hg levels and 127 were negatively correlated (Supplementary Table 1). Figure 7 shows a heat map of the ten highest positive and ten lowest negative partial correlation coefficients with Hg levels in the AU group compared to their partial correlation coefficients in the TD group. The most represented molecular and cellular functions for the 189 probe sets (Table 2) were cell morphology (DIRAS1, PIP5K3, ANK1, CAPZB, ASAP1, RRBP1, P-value range 1.64E−04 to 4.39E−02), amino acid metabolism (GGT1, SLC25A22, SLC7A8, P-value range 7.33E−03 to 3.56E−02), antigen presentation (PPT2, NLRP1, NOD1, SLC11A1, HLA-DQA1, and HLA-DQB1, P-value range 7.44E−03 to 1.77E−02), carbohydrate metabolism (PIP5K3, SLC35B4, P-value range 7.44E−03 to 2.95E−02), and cell cycle (SASS6, PIP5K3, AMACR, P-value range 7.44E−03 to 2.22E−02). Analysis of the promoter sequences of these genes using Genomatix software revealed binding site sequences for the transcription factor KLF family (P = 0.00015, minimum 86% of sequences). Chromosomes 1 and 8 showed significant enrichment with 25 and 11 out of the 189 probe sets, respectively (Table 2, Supplementary Fig. 3). Functional annotation of the genes on chromosome 1 showed that they are involved in heat shock protein binding (8%, P = 0.07), whereas the genes on chromosome 8 are involved in protein kinase activity (11%, P = 0.1). Cytoband enrichment was noted for 1p33 (3/189, P = 0.02) and 6p21.3 (5/189, P = 0.05).
Genes with the ten highest positive and the ten lowest negative correlations of gene expression with Hg levels for the AU subjects (List D). Heat map of the partial correlation coefficients (numbers in boxes) of the gene expression and Hg levels in TD and AU. Genes are on the Y-axis, and diagnoses are on the X-axis
Analysis of All Genes Correlating with Hg Levels in AU and/or TD
Finally, we performed an analysis of all genes whose expression correlated with Hg levels regardless of diagnostic group or sign of correlation (all four gene lists combined—A–D). The rationale for doing this was to ask whether there were genes in all four lists that were associated with the same functional networks. The IPA analysis revealed that the top molecular and cellular functions of the genes correlating with Hg levels were cell morphology (29 molecules, P-value range 1.19E−03 to 4.01E−02), cell death (24 molecules, P-value range 2.35E−03 to 4.60E−02), cellular assembly and organization (25 molecules, P-value range 3.87E−03 to 4.01E−02), cellular movement (8 molecules, P-value range 3.87E−03 to 4.988E−02), and cellular development (15 molecules, P-value range 5.72E−03 to 4.49E−02). The top disease and disorder biofunctions correlating with Hg levels were cancer (30 molecules, P-value range 6.03E−04 to 4.98E−02), hematological disease (14 molecules, P-value range 6.02E−04 to 4.89E−02), genetic disorder (185 molecules, P-value range 1.81E−03 to 3.98E−02), neurological disease (103 molecules, P-value range 2.35E−03 to 3.98E−02), and skeletal and muscular disorders (11 molecules, P-value range 1.04E−02 to 3.98E−02). The top canonical pathways correlating with Hg levels were GABA receptor signaling (4/55 molecules, P-value 1.16E−02), lysine degradation (7/234 molecules, P-value 2.74E−02), thyroid cancer signaling (3/45 molecules, P-value 4.04E−02), and cyanoamino acid metabolism (2/64 molecules, P-value 4.98E−02). Figure 8 shows the IPA network with the highest score. It is notable that many genes are regulated in this network, but that the Hsp70, Hsp90, proteasome, NFκB, Raf, and caspase nodes themselves are not regulated.
Comparison of Genes Correlating with Hg Levels Only in TD (List C) and Only in AU (List D)
To gain more insight into the associations between transcript levels and circulating Hg levels in the TD and the AU children, and to evaluate the biological significance of the differences between the TD and AU boys, we compared the genes that correlated with Hg levels only in the TD group (List C), and genes that correlated with Hg levels only in the AU group (List D). We split each gene list into two lists, based on the direction of the correlation coefficients—into a list of genes whose expression positively correlates with Hg levels and into a list of genes whose expression negatively correlates with Hg levels. The IPA canonical pathways over-represented in at least one of the four categories (TD (+) and TD (−) from List C, and AU (+) and AU (−) from List D, signs represent the direction of the partial correlation) are shown in Fig. 9. The TGF-beta signaling pathway (Fig. 10) and molecular mechanisms of cancer (P = 3.48E−02, 2/86 and P = 3.66E−02, 4/370, respectively) are over-represented in the positively correlating genes between Hg level and gene expression in the TD group. Lysine degradation (P = 3.63E−02, 4/234) was over-represented in the negatively correlating genes between Hg level and gene expression in the TD group. p38 MAPK signaling and α-adrenergic signaling were over-represented in the positively correlating genes between Hg level and gene expression in the AU group.
Canonical pathways over-represented in at least one of the four categories from the genes that correlate with mercury levels in TD subjects (List C) and that correlate with mercury levels in AU subjects (List D). TD (+) = red = positively correlated genes from List C. TD (−) = blue = negatively correlated genes from List C. AU (+) = hatched red = positively correlated genes from List D. AU (−) = hatched blue = negatively correlated genes from List D
Discussion
Though some of the genes and pathways are similar, the major finding of the study is that almost all of the genes that correlate with circulating Hg levels in AU boys are different from TD boys, and almost all of the genes that correlate with circulating Hg levels in TD boys are different from AU boys. These transcriptional differences were observed even though the Hg levels were low and not significantly different between the groups in our study. Since the data reported are only correlations, no conclusions can be drawn as to whether Hg plays any role in the pathogenesis of autism based upon the current results. Given the low Hg levels and the fact that this is a cross-sectional study, no cause and effect relationship should be drawn from the current data. Though the data can be interpreted in several ways, we suggest that the different transcriptional programs associated with Hg in AU compared to TD subjects may be related to the genetic differences in the two groups of children.
Common Genes (List A and List B) and Common Pathways
The major finding of this study is that very few genes that correlated with Hg levels in AU subjects also correlated with Hg levels in TD subjects. There were only 15 genes that correlated with Hg levels in both the TD and AU groups—and that were not significantly different between the two groups. These genes were involved in apoptosis, the immune response, and response to oxidative stress. Moreover, there were only 11 genes whose expression correlated inversely with Hg levels in AU compared to TD subjects—that is genes correlating with Hg levels in both groups but in opposite directions. These genes were involved in pathways contributing to neuronal development and neuronal survival and cell death. Notably, one gene (FLNB, filamin B, beta (actin binding protein 278)), which was positively correlated with Hg levels in TD and negatively correlated in AU subjects, is involved in anchoring and biogenesis of cytoskeleton and organization of actin filaments (Xu et al. 1998; Tu et al. 2003; Leedman et al. 1993). A number of studies show that most heavy metals, including zinc, arsenic, mercury, and cadmium, can disrupt actin and microtubules in intact cells (Templeton 2000).
The analysis of all genes correlating with Hg levels in all AU and TD subjects showed a number of common networks. Figure 8 is a good example where different genes from the different groups are regulated within the same network. This could suggest that similar functions might be accomplished with different genes in AU compared to TD, and that homeostasis is maintained in spite of the genetic differences of AU and TD subjects.
Genes Correlating with Hg Levels in TD but not in AU Boys (List C)
Thus, most of the genes whose expression correlated with Hg levels in the TD group were different from those whose expression correlated with Hg levels in the AU group. For the TD subjects, the top biological function for the genes that correlated with Hg levels was cell death. Some of these genes were involved in apoptosis of neurons (NFATC4, BAD, BDNF) (Graef et al. 2003; Ottilie et al. 1997; Hoey et al. 1995; Botta et al. 2004; Kerschensteiner et al. 1999; Mamounas et al. 1995), survival of neurons, recovering and regeneration of axons (BDNF, RTN4) (Kerschensteiner et al. 1999; GrandPre et al. 2000), apoptosis of pre-T lymphocytes (BCL2L11) (Bouillet et al. 1999), survival of pro-B lymphocytes, and survival of memory B cells (BRAF, BCL2L11, BDNF, TCF3) (Oliver et al. 2004; Fischer et al. 2007). It is interesting that most of these correlations are negative. Chronic exposure to low concentrations of heavy metals, including mercury, causes immune dysfunction related in large part to apoptosis of leukocytes (Pollard and Hultman 1997; Zelikoff and Gardner 1996; Laiosa et al. 2007; Madureira et al. 2007; Ziemba et al. 2005; Mondal et al. 2005; McCabe et al. 2003, 2005; Cunha et al. 2004; Sarmento et al. 2004; Kuo and Lin-Shiau 2004; Colombo et al. 2004; Field et al. 2003). Both inorganic and organic mercurials cause human T-cell apoptosis with mitochondria being the target organelle for the induction of cell death (Shenker et al. 1999). In addition, microarray analysis of Hg-induced changes in gene expression in human liver carcinoma (HepG2) cells revealed genes involved in apoptosis and metabolic regulation with significant effects on genes involved in immune system pathways (Ayensu and Tchounwou 2006). However, the levels of Hg applied to the cells were much higher than the Hg levels of the subjects in our study.
The second major biological function for genes that correlated with Hg levels in TD subjects was cell morphology and cell death. Mercury-induced cell death can be apoptotic or nonapoptotic. The nonapoptotic cell death produced by mercury is associated with a specific cellular morphology (Pollard et al. 1997, 2000). Mercury-induced apoptosis results from dys-regulated signal transduction that leads to inactivation of enzymes, generation of free radicals and peroxides, protein conformation changes, and inhibition of transport processes which disrupt cell membrane permeability (Tchounwou et al. 2003; Clarkson and Magos 2006; Counter and Buchanan 2004; Zahir et al. 2005). Genes associated with transmembrane potential and permeabilization of mitochondria correlated with Hg levels in TD subjects in our study. Mercury-induced membrane changes of mitochondria associated with glutathione imbalance and antioxidant defenses have been reported in TD versus AU transformed lymphoblastoid lines (James et al. 2009). Mercury increases mitochondrial permeability, which is characterized both by a decline in transmembrane potential and intracellular pH, as well as the generation of reactive oxygen species and a decline in T-cell GSH content (Shenker et al. 2000). In our study, Granulysin (GNLY) showed a positive correlation with Hg levels in the TD group. GNLY is a cytolytic and pro-inflammatory molecule, which contributes to mitochondrial damage (Krensky and Clayberger 2009). B-cell lymphoma 2-like protein 11 (apoptosis facilitator) (BCL2L11) showed a negative correlation with Hg in the TD group in our study. BCL2L11 (also known as BIM) is a BH3-only protein from the Bcl-2 family (Youle and Strasser 2008), whose family members are the main regulators of programmed cell death via the mitochondrial (intrinsic) apoptotic pathway. Interactions between pro- and anti-apoptotic proteins of the Bcl-2 family decide the fate of cells in response to stress signals. BCL2L11 is essential for hematopoietic homeostasis and thymocyte negative selection, and suppresses autoimmunity (Youle and Strasser 2008).
Some of the cell morphology genes that correlated with Hg levels in TD subjects also have functions within the central nervous system. These included genes that affect neuronal survival (ABL1 (+), BDNF (−)), sprouting and projection of axons (BDNF (−), RTN4 (−)), plasticity of the synapse (BDNF (−), NFATC4 (+), PMCH (−)), and remodeling of axons (SFRP1 (−)) (Kerschensteiner et al. 1999; GrandPre et al. 2000; Gascon et al. 2007; DeFreitas et al. 2001; Mamounas et al. 2000; Fournier et al. 2001; Seil 1998; Groth and Mermelstein 2003; Krylova et al. 2002). It is possible that mercury affects these genes in the brain as well as the blood, and this might relate to neurotoxicity observed following exposure to much higher levels of mercury (Rice 2008; Guzzi and La Porta 2008).
We observed a negative correlation between GCLM and Hg levels in TD, but no correlation in AU. GCLM is a member of the glutamate-cysteine ligase complex, which is the first rate-limiting enzyme of the glutathione synthesis, and is the major route of mercury detoxification.
James et al. (2009) observed a reduced glutathione reserve capacity in both cytosol and mitochondria in lymphoblastoid cells derived from children with autism when compared to children without autism. Glutathione reserve capacity is an indicator of systemic redox status that can be used to assess and treat individuals at risk of oxidative stress-related pathology (Jones 2006a, b). In our study, we observed a negative partial correlation between GCLM transcript abundance and Hg levels in the TD group, while there was no significant correlation in the AU group. GCLM is transcriptionally regulated in human HepG2 cells (Lee et al. 2006). We also observed a negative partial correlation between GGT1 transcript abundance and Hg levels in the AU group, while there was no significant correlation in the TD group (see section “Genes Whose Expression Correlates with Hg Levels in AU but not in TD Boys (List D)”). GGT1 is involved in the utilization of l-glutamate, l-cysteine, and glutathione metabolism. It is involved in the generation of l-glutamate, which is a substrate for glutathione synthesis. Thus, if there is negative regulation of this gene, there may be less substrate for the glutathione biosynthesis pathway, which may affect the redox status. Overall, our studies of transcript abundance in blood of AU and TD boys are difficult to relate to the in vitro studies of AU and TD lymphoblastoid cell lines except to say that both approaches show associations between genes in the glutathione pathways and Hg levels.
CDKN2AIPNL and ZMYM6 negatively correlated with Hg levels in this study and both were downregulated with mercury treatment in human liver carcinoma (HepG2) cells (Ayensu and Tchounwou 2006). ALDH3B1, RPS9, SUB1, and VPS25 also were negatively correlated with Hg in TD subjects in this study and were downregulated in Hg-treated yeast (Jin et al. 2008).
Genes whose expression correlated similarly with Hg levels both in TD and AU (List A) and whose expression correlated with Hg levels only in the AU group (List D) have transcription factor (TF) binding sites for SP1 and Kruppel-like factor (KLF)-like TFs, respectively. KLF6, which is an Sp1-like zinc finger transcription factor, is a transactivator of inducible nitric oxide synthase (iNOS) (Warke et al. 2003), which produces NO and modulates inflammatory signals and oxidative stress (Mungrue et al. 2003).
Genes Whose Expression Correlates with Hg Levels in AU but not in TD Boys (List D)
One of the top biological functions in the list of genes that correlated with Hg levels in AU but not TD boys was amino acid metabolism. The genes included gamma-glutamyltransferase (GGT1/2/3; probe 244179_x_at); as well as solute carrier family 25 (mitochondrial carrier: glutamate), member 22 (SLC25A22 (−)), and solute carrier family 7 (cationic amino acid transporter, y+ system), member 8 (SLC7A8 (−)) which are involved in the transport of l-amino acids (Pineda et al. 1999). SLC7A8, which had negative partial correlation with Hg levels in the AU group in our study, was upregulated in mercury-treated yeast cells (Jin et al. 2008).
FAF1 (Fas-associated factor 1) expression was positively correlated with Hg levels in AU but not TD subjects. Fas is a member of the tumor necrosis receptor factor family, which can induce apoptosis when activated by Fas-ligand binding (Chu et al. 1995). Apoptosis induced by Fas plays an important role in the development and function of the immune system (Lowin et al. 1994; Nagata 1994). Genetic studies in mice show that defects in Fas-mediated apoptosis resulted in abnormal development and function in the immune system (Takahashi et al. 1994). FAF-1 also inhibits the NF-κB activation induced by various stimuli, including tumor necrosis factor-α, interleukin-1β, and lipopolysaccharide (Park et al. 2004). NF-κB is a central regulator of genes involved in the immune response, inflammatory response, and apoptosis (Beg and Baltimore 1996; Pahl 1999; Baldwin 1996).
CHCHD4 (negatively correlated with Hg level in AU), ELOVL1, SLC7A8, and Cxorf34 (positively correlated with Hg level in AU) responded to mercury in yeast in the same direction as in our AU subjects. NTHL1 (positively correlated with Hg level in AU) was downregulated in yeast after mercury treatment, whereas MYBBP1A (positively correlated in AU) was up- or downregulated depending on the treatment dose (Jin et al. 2008).
HLA-DQ, an αβ heterodimer of the MHC Class II type which is a cell surface molecule found on antigen presenting cells, presents antigens to T-lymphocytes (Germain and Margulies 1993; Janeway et al. 1997; Kwok and Nepom 1998). The α and β chains are encoded by HLA-DQA1 and HLA-DQB1, respectively, and both the α- and β-chains vary greatly in the population. Different DQ isoforms can bind to and present different antigens to T-cells. Several lines of evidence suggest that mercury toxicity depends on the mercury species, dosage, individuality, and the type of MHC association (Ayensu et al. 2004; Hanley et al. 1997, 2002). Moreover, chronic exposure to low concentrations of heavy metals such as mercury results in immune dysfunction (Ayensu et al. 2004; Hanley et al. 1997, 2002). Immune dysregulation has been observed in autism, and has been postulated to play a role in the development and pathogenesis of autism in some cases (Warren et al. 1996, 1997; Singh and Rivas 2004; Ashwood et al. 2006; Gupta et al. 1996). In this study, we found that HLA-DQA1 was negatively correlated, whereas HLA-DQB1 was positively correlated with Hg levels in AU but not TD subjects. They both were downregulated in response to mercury in human liver carcinoma (HepG2) cells (Ayensu and Tchounwou 2006).
Comparison of Genes Correlating with Hg Levels Only in TD or Only in AU (List C or D)
There is a different association between transcript level and circulating Hg levels in the TD and AU subjects that involves genes from the TGF-β signaling pathway. TGF-β signaling regulates cell growth, cell differentiation, cellular homeostasis, apoptosis, and immune responses. TGF-β binds to and activates the TGF-β receptor, which then activates the SMAD family of transcriptional regulators. SMAD2 and SMAD3 mediate TGF-β and activin signals, while SMAD1, SMAD5, and SMAD8 act downstream of bone morphogenetic proteins (BMPs) (Zhang and Derynck 1999; ten Dijke et al. 2000; Massague and Wotton 2000). SMADs dimerize with SMAD4 and translocate to the nucleus where the complex modulates transcription of target genes. In our study, we found bone morphogenetic protein receptor type 2 (serine/threonine kinase) (BMPR2) and SMAD2 to be positively correlated with Hg levels in TD but not in AU subjects. Notably, Ashwood et al. (2008) observed decreased TGF-β1 plasma levels in children with autism spectrum disorders when compared to typically developing controls.
Hg Levels in AU and TD
Hg levels in children from the CHARGE study with and without autism have been studied in detail (Hertz-Picciotto et al. 2009). The Hg levels in the TD and AU groups were quite similar in this sample to those in the larger CHARGE sample: means of 0.60 μg/l for TDs in both groups, and means of 0.46 and 0.49 μg/l for the AU/ASD children in this subsample and the larger study (Hertz-Picciotto et al. 2009). Thus, the sample of subjects in this study is representative of the CHARGE study as a whole. None of the samples used in this study had Hg levels higher than the EPA reference level of 5.8 μg/l. The median Hg concentration and inter-quartile range were comparable to those of the U.S. population of children ages 1–5 years in 1999–2002 (McDowell et al. 2004). It is notable, therefore, that the gene expression differences described here occurred in the EPA allowable range. This is consistent with previously reported dose-dependent effects of Hg (Tchounwou et al. 2003; Clarkson and Magos 2006; Counter and Buchanan 2004; Zahir et al. 2005) and with a dose-dependent transcriptional response in human liver carcinoma cell lines treated with mercury (Ayensu and Tchounwou 2006). It is important to emphasize that acute intoxication with much higher mercury concentrations or even chronic exposure to much higher Hg concentrations might produce different transcript associations.
Limitations of the Study
Samples collected from children ages 2–5 in this study do not directly address the role of Hg as a possible causal factor for autism which likely begins in utero or shortly after birth. In addition, it is important to emphasize that one cannot infer cause and effect from the data presented in this study. Our data shows that there are associations between transcript levels and circulating Hg levels which are different in the AU compared to the TD group. It is possible that these low levels of Hg cause changes of gene expression in the white blood cells that could affect the function of immune cells that could play a role in the pathogenesis of autism. Alternatively, and perhaps more likely, the altered transcriptomic associations with low levels of Hg in AU may have no functional effect on immune cells, brain, or other organs. Further studies on protein levels of the genes whose expression was associated with mercury levels should be conducted. Protein levels may or may not reflect RNA levels, particularly in stressed cells. Again, correlation does not mean causality and extreme caution should be exercised when interpreting the correlations reported here. In addition, since this is a one-time study, the correlation of an individual gene might not be replicated in future studies due to stochasticity.
The P-values which were used to derive the gene lists indicate statistical but not biological significance. It is unclear what level of correlation of gene expression and Hg levels represents biological significance. Correlation is also affected by “outlying” values. One way of reducing this effect is by log-transforming the data as was done here. Also, the somewhat unequal sample sizes may affect the calculation of the P-values and thus lead to some skewing of the data. The strengths of the correlations were not large for most of the identified genes. Therefore, caution should be exercised regarding the correlation of gene expression with Hg level for a given gene.
No gene reported here passed a multiple comparison correction for statistical significance. We addressed this problem in part by focusing on genes common to at least two of the ANCOVAs. However, the finding that so few genes correlated with Hg levels in both AU and TD subjects implies that multiple comparisons had no impact on the major conclusion of this study: AU and TD children have different associations between transcript level and circulating Hg levels. Increasing multiple comparisons would have made it likely that there would be many transcripts that correlated with Hg and were common to the AU and TD groups.
This study evaluated a single sample of blood where only 5% of the body burden of Hg is found (Burbacher et al. 2005; Stinson et al. 1989). The different Hg species were not measured (Goldman and Shannon 2001; Clarkson 2002). It is likely that different Hg species induce specific genes and affect specific enzymes that mediate Hg species specific toxicity (Clarkson and Magos 2006; Aschner et al. 2006).
The observed correlation between Hg levels and gene expression might be due to other factors that were not included in our ANCOVA models. Different diets of AU compared to TD subjects might affect gene expression. For example, fish is a major source of organic mercury intake and in the larger CHARGE study sample (Hertz-Picciotto et al. 2009) accounted for the differences in mercury levels between subjects. Fish contain n-3 polyunsaturated fatty acids, which are potent regulators of gene expression (Price et al. 2000). It is possible that some of the associations observed in this study are confounded by n-3 polyunsaturated fatty acids levels or other molecules found in fish.
Abbreviations
- ABL1:
-
c-abl oncogene 1, receptor tyrosine kinase
- ALDH3B1:
-
Aldehyde dehydrogenase 3 family, member B1
- ALS2:
-
Amyotrophic lateral sclerosis 2 (juvenile)
- AP2A2:
-
Adaptor-related protein complex 2, alpha 2 subunit
- BAD:
-
BCL2-antagonist of cell death
- BCL2L11:
-
BCL2-like 11 (apoptosis facilitator)
- BDNF:
-
Brain-derived neurotrophic factor
- BMPR2:
-
Serine/threonine receptor kinase
- BRAF:
-
V-raf murine sarcoma viral oncogene homolog B1
- CADM1:
-
Cell adhesion molecule 1
- CDKN2AIPNL:
-
CDKN2A interacting protein N terminal like
- CHCHD4:
-
Coiled-coil-helix-coiled-coil-helix domain containing 4
- CXorf34:
-
Chromosome X open reading frame 34
- ELOVL1:
-
Elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 1
- FEZ2:
-
Fasciculation and elongation protein zeta 2 (zygin II)
- FLNB:
-
Filamin B, beta (actin binding protein 278)
- GCLM:
-
Glutamate-cysteine ligase, modifier subunit
- GGT:
-
Gamma-glutamyltransferase
- GNLY:
-
Granulysin
- GPCRs:
-
G Protein-coupled receptors
- HLA-DQA1:
-
Major histocompatibility complex, class II, DQ alpha 1
- HLA-DQB1:
-
major histocompatibility complex, class II, DQ beta 1
- KRT1:
-
Keratin 1 (epidermolytic hyperkeratosis)
- LMLN:
-
Leishmanolysin-like (metallopeptidase M8 family)
- LOC100129026:
-
Hypothetical protein LOC100129026
- MAPK:
-
Mitogen-activated protein kinases; serine/threonine receptor kinases
- MSH6:
-
mutS homolog 6 (E. coli)
- MUC1:
-
Mucin 1, cell surface associated
- MYBBP1A:
-
MYB binding protein (P160) 1a
- NCAPD3:
-
Non-SMC condensin II complex, subunit D3
- NFATC4:
-
Nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4
- NTHL1:
-
nth endonuclease III-like 1 (E. coli)
- NTRK3:
-
Neurotrophic tyrosine kinase, receptor, type 3
- PMCH:
-
Pro-melanin-concentrating hormone
- PXDN:
-
Peroxidasin homolog (Drosophila)
- RPS9:
-
Ribosomal protein S9
- RTN4:
-
Reticulon 4
- SCARB1:
-
Scavenger receptor class B, member 1
- SFRP1:
-
Secreted frizzled-related protein 1
- SLC25A22:
-
Solute carrier family 25 (mitochondrial carrier: glutamate), member 22
- SLC7A8:
-
Solute carrier family 7 (cationic amino acid transporter, y+ system), member 8
- SMAD2:
-
SMAD family member 2
- SSTR3:
-
Somatostatin receptor 3
- SUB1:
-
SUB1 homolog (S. cerevisiae)
- TCF3:
-
T-cell factor; transcription factor
- TXNDC17:
-
Thioredoxin domain containing 17
- VPS25:
-
Vacuolar protein sorting 25 homolog (S. cerevisiae)
- ZBTB16:
-
Zinc finger and BTB domain containing 16
- ZMYM6:
-
Zinc finger, MYM-type 6
References
Aschner M, Syversen T, Souza DO, Rocha JB (2006) Metallothioneins: mercury species-specific induction and their potential role in attenuating neurotoxicity. Exp Biol Med (Maywood) 231(9):1468–1473
Ashwood P, Wills S, Van de Water J (2006) The immune response in autism: a new frontier for autism research. J Leukoc Biol 80(1):1–15
Ashwood P, Enstrom A, Krakowiak P, Hertz-Picciotto I, Hansen RL, Croen LA et al (2008) Decreased transforming growth factor beta1 in autism: a potential link between immune dysregulation and impairment in clinical behavioral outcomes. J Neuroimmunol 204(1–2):149–153
Ayensu WK, Tchounwou PB (2006) Microarray analysis of mercury-induced changes in gene expression in human liver carcinoma (HepG2) cells: importance in immune responses. Int J Environ Res Public Health 3(2):141–173
Ayensu WK, Tchounwou PB, McMurray RW (2004) Molecular and cellular mechanisms associated with autoimmune diseases. Int J Environ Res Public Health 1(1):39–73
Baldwin AS Jr (1996) The NF-kappa B and I kappa B proteins: new discoveries and insights. Annu Rev Immunol 14:649–683
Beg AA, Baltimore D (1996) An essential role for NF-kappaB in preventing TNF-alpha-induced cell death. Science 274(5288):782–784
Botta D, Franklin CC, White CC, Krejsa CM, Dabrowski MJ, Pierce RH et al (2004) Glutamate-cysteine ligase attenuates TNF-induced mitochondrial injury and apoptosis. Free Radic Biol Med 37(5):632–642
Bouillet P, Metcalf D, Huang DC, Tarlinton DM, Kay TW, Kontgen F et al (1999) Proapoptotic Bcl-2 relative Bim required for certain apoptotic responses, leukocyte homeostasis, and to preclude autoimmunity. Science 286(5445):1735–1738
Burbacher TM, Shen DD, Liberato N, Grant KS, Cernichiari E, Clarkson T (2005) Comparison of blood and brain mercury levels in infant monkeys exposed to methylmercury or vaccines containing thimerosal. Environ Health Perspect 113(8):1015–1021
Chu K, Niu X, Williams LT (1995) A Fas-associated protein factor, FAF1, potentiates Fas-mediated apoptosis. Proc Natl Acad Sci USA 92(25):11894–11898
Clarkson TW (2002) The three modern faces of mercury. Environ Health Perspect 110(Suppl 1):11–23
Clarkson TW, Magos L (2006) The toxicology of mercury and its chemical compounds. Crit Rev Toxicol 36(8):609–662
Colombo M, Hamelin C, Kouassi E, Fournier M, Bernier J (2004) Differential effects of mercury, lead, and cadmium on IL-2 production by Jurkat T cells. Clin Immunol 111(3):311–322
Counter SA, Buchanan LH (2004) Mercury exposure in children: a review. Toxicol Appl Pharmacol 198(2):209–230
Cunha EM, Oliveira MJ, Ferreira PG, Aguas AP (2004) Mercury intake by inflammatory phagocytes: in vivo cytology of mouse macrophages and neutrophils by X-ray elemental microanalysis coupled with scanning electron microscopy. Hum Exp Toxicol 23(9):447–453
DeFreitas MF, McQuillen PS, Shatz CJ (2001) A novel p75NTR signaling pathway promotes survival, not death, of immunopurified neocortical subplate neurons. J Neurosci 21(14):5121–5129
Eisenhart C (1947) The assumptions underlying the analysis of variance. Biometrics 3:20
Ercal N, Gurer-Orhan H, Aykin-Burns N (2001) Toxic metals and oxidative stress part I: mechanisms involved in metal-induced oxidative damage. Curr Top Med Chem 1(6):529–539
Field AC, Bloch MF, Bellon B (2003) Neonatal tolerance to a Th2-mediated autoimmune disease generates CD8+ Tc1 regulatory cells. J Autoimmun 21(3):201–212
Fischer SF, Bouillet P, O’Donnell K, Light A, Tarlinton DM, Strasser A (2007) Proapoptotic BH3-only protein Bim is essential for developmentally programmed death of germinal center-derived memory B cells and antibody-forming cells. Blood 110(12):3978–3984
Fournier AE, GrandPre T, Strittmatter SM (2001) Identification of a receptor mediating Nogo-66 inhibition of axonal regeneration. Nature 409(6818):341–346
Gascon E, Vutskits L, Jenny B, Durbec P, Kiss JZ (2007) PSA-NCAM in postnatally generated immature neurons of the olfactory bulb: a crucial role in regulating p75 expression and cell survival. Development 134(6):1181–1190
Germain RN, Margulies DH (1993) The biochemistry and cell biology of antigen processing and presentation. Annu Rev Immunol 11:403–450
Goldman LR, Shannon MW (2001) Technical report: mercury in the environment: implications for pediatricians. Pediatrics 108(1):197–205
Graef IA, Wang F, Charron F, Chen L, Neilson J, Tessier-Lavigne M et al (2003) Neurotrophins and netrins require calcineurin/NFAT signaling to stimulate outgrowth of embryonic axons. Cell 113(5):657–670
GrandPre T, Nakamura F, Vartanian T, Strittmatter SM (2000) Identification of the Nogo inhibitor of axon regeneration as a Reticulon protein. Nature 403(6768):439–444
Groth RD, Mermelstein PG (2003) Brain-derived neurotrophic factor activation of NFAT (nuclear factor of activated T-cells)-dependent transcription: a role for the transcription factor NFATc4 in neurotrophin-mediated gene expression. J Neurosci 23(22):8125–8134
Gupta S, Aggarwal S, Heads C (1996) Dysregulated immune system in children with autism: beneficial effects of intravenous immune globulin on autistic characteristics. J Autism Dev Disord 26(4):439–452
Guzzi G, La Porta CA (2008) Molecular mechanisms triggered by mercury. Toxicology 244(1):1–12
Hanley GA, Schiffenbauer J, Sobel ES (1997) Class II haplotype differentially regulates immune response in HgCl2-treated mice. Clin Immunol Immunopathol 84(3):328–337
Hanley GA, Schiffenbauer J, Sobel ES (2002) Resistance to HgCl2-induced autoimmunity in haplotype-heterozygous mice is an intrinsic property of B cells. J Immunol 161:1778–1789
Hertz-Picciotto I, Croen LA, Hansen R, Jones CR, van de Water J, Pessah IN (2006) The CHARGE study: an epidemiologic investigation of genetic and environmental factors contributing to autism. Environ Health Perspect 114(7):1119–1125
Hertz-Picciotto I, Green PG, Delwiche L, Hansen R, Walker C, Pessah IN (2009) Blood mercury concentrations in CHARGE study children with and without autism. Environ Health Perspect. doi:10.1289/ehp.0900736
Hoey T, Sun YL, Williamson K, Xu X (1995) Isolation of two new members of the NF-AT gene family and functional characterization of the NF-AT proteins. Immunity 2(5):461–472
James SJ, Rose S, Melnyk S, Jernigan S, Blossom S, Pavliv O et al (2009) Cellular and mitochondrial glutathione redox imbalance in lymphoblastoid cells derived from children with autism. FASEB J 23(8):2374–2383
Janeway CA, Travers P, Hunt S, et al. (eds) (1997) Antigen recognition by T-lymphocytes. In: Immunobiology: the immune system in health and disease, 3rd edn. Garland Publishing, New York
Jin YH, Dunlap PE, McBride SJ, Al-Refai H, Bushel PR, Freedman JH (2008) Global transcriptome and deletome profiles of yeast exposed to transition metals. PLoS Genet 4(4):e1000053
Jones DP (2006a) Extracellular redox state: refining the definition of oxidative stress in aging. Rejuvenation Res 9(2):169–181
Jones DP (2006b) Redefining oxidative stress. Antioxid Redox Signal 8(9–10):1865–1879
Kerschensteiner M, Gallmeier E, Behrens L, Leal VV, Misgeld T, Klinkert WE et al (1999) Activated human T cells, B cells, and monocytes produce brain-derived neurotrophic factor in vitro and in inflammatory brain lesions: a neuroprotective role of inflammation? J Exp Med 189(5):865–870
Krensky AM, Clayberger C (2009) Biology and clinical relevance of granulysin. Tissue Antigens 73(3):193–198
Krylova O, Herreros J, Cleverley KE, Ehler E, Henriquez JP, Hughes SM et al (2002) WNT-3, expressed by motoneurons, regulates terminal arborization of neurotrophin-3-responsive spinal sensory neurons. Neuron 35(6):1043–1056
Kuo TC, Lin-Shiau SY (2004) Early acute necrosis and delayed apoptosis induced by methyl mercury in murine peritoneal neutrophils. Basic Clin Pharmacol Toxicol 94(6):274–281
Kwok WW, Nepom GT (eds) (1998) Genetic influences: major histocompatibility complex. In: The autoimmune diseases, 3rd edn. Academic Press, New York
Laiosa MD, Eckles KG, Langdon M, Rosenspire AJ, McCabe MJ Jr (2007) Exposure to inorganic mercury in vivo attenuates extrinsic apoptotic signaling in Staphylococcal aureus enterotoxin B stimulated T-cells. Toxicol Appl Pharmacol 225(3):238–250
Le Couteur A, Lord C, Rutter M (2003) Autism Diagnostic Interview-Revised (ADI-R). Western Psychological Services, Los Angeles
Lee JI, Kang J, Stipanuk MH (2006) Differential regulation of glutamate-cysteine ligase subunit expression and increased holoenzyme formation in response to cysteine deprivation. Biochem J 393(Pt 1):181–190
Leedman PJ, Faulkner-Jones B, Cram DS, Harrison PJ, West J, O’Brien E et al (1993) Cloning from the thyroid of a protein related to actin binding protein that is recognized by Graves disease immunoglobulins. Proc Natl Acad Sci USA 90(13):5994–5998
London EA (2000) The environment as an etiologic factor in autism: a new direction for research. Environ Health Perspect 108(Suppl 3):401–404
Lord C, Rutter M, Le Couteur A (1994) Autism Diagnostic Interview-Revised: a revised version of a diagnostic interview for caregivers of individuals with possible pervasive developmental disorders. J Autism Dev Disord 24(5):659–685
Lord C, Pickles A, McLennan J, Rutter M, Bregman J, Folstein S et al (1997) Diagnosing autism: analyses of data from the Autism Diagnostic Interview. J Autism Dev Disord 27(5):501–517
Lord C, Rutter M, DiLavore PC, Risi S (2000) The Autism Diagnostic Observation Schedule (ADOS). Western Psychological Services, Los Angeles
Lord C, Rutter M, DiLavore PC, Risi S (2003) The autism diagnostic observation schedule manual. Western Psychological Services, Los Angeles
Lowin B, Hahne M, Mattmann C, Tschopp J (1994) Cytolytic T-cell cytotoxicity is mediated through perforin and Fas lytic pathways. Nature 370(6491):650–652
Madureira P, Cunha EM, Aguas AP (2007) Acute depletion and recovery of peritoneal B-1 lymphocytes in BALB/c mice after a single injection of mercury chloride. Immunopharmacol Immunotoxicol 29(2):311–322
Mamounas LA, Blue ME, Siuciak JA, Altar CA (1995) Brain derived neurotrophic factor promotes the survival and sprouting of sertonergic. J Neurosci 15:7929–7939
Mamounas LA, Altar CA, Blue ME, Kaplan DR, Tessarollo L, Lyons WE (2000) BDNF promotes the regenerative sprouting, but not survival, of injured serotonergic axons in the adult rat brain. J Neurosci 20(2):771–782
Massague J, Wotton D (2000) Transcriptional control by the TGF-beta/Smad signaling system. EMBO J 19(8):1745–1754
McCabe MJ Jr, Whitekus MJ, Hyun J, Eckles KG, McCollum G, Rosenspire AJ (2003) Inorganic mercury attenuates CD95-mediated apoptosis by interfering with formation of the death inducing signaling complex. Toxicol Appl Pharmacol 190(2):146–156
McCabe MJ Jr, Eckles KG, Langdon M, Clarkson TW, Whitekus MJ, Rosenspire AJ (2005) Attenuation of CD95-induced apoptosis by inorganic mercury: caspase-3 is not a direct target of low levels of Hg2+. Toxicol Lett 155(1):161–170
McCarty LS, Borgert CJ (2006a) Review of the toxicity of chemical mixtures: theory, policy, and regulatory practice. Regul Toxicol Pharmacol 45(2):119–143
McCarty LS, Borgert CJ (2006b) Review of the toxicity of chemical mixtures containing at least one organochlorine. Regul Toxicol Pharmacol 45(2):104–118
McDowell MA, Dillon CF, Osterloh J, Bolger PM, Pellizzari E, Fernando R et al (2004) Hair mercury levels in U.S. children and women of childbearing age: reference range data from NHANES 1999–2000. Environ Health Perspect 112(11):1165–1171
Mokrzan EM, Kerper LE, Ballatori N, Clarkson TW (1995) Methylmercury-thiol uptake into cultured brain capillary endothelial cells on amino acid system L. J Pharmacol Exp Ther 272(3):1277–1284
Mondal TK, Li D, Swami K, Dean JK, Hauer C, Lawrence DA (2005) Mercury impairment of mouse thymocyte survival in vitro: involvement of cellular thiols. J Toxicol Environ Health A 68(7):535–556
Morrison DF (ed) (1976) Multivariate statistical methods, 2nd edn. McGraw-Hill, New York
Mungrue IN, Bredt DS, Stewart DJ, Husain M (2003) From molecules to mammals: what’s NOS got to do with it? Acta Physiol Scand 179(2):123–135
Nagata S (1994) Mutations in the Fas antigen gene in lpr mice. Semin Immunol 6(1):3–8
Newland MC, Paletz EM, Reed MN (2008) Methylmercury and nutrition: adult effects of fetal exposure in experimental models. Neurotoxicology 29(5):783–801
Oliver PM, Wang M, Zhu Y, White J, Kappler J, Marrack P (2004) Loss of Bim allows precursor B cell survival but not precursor B cell differentiation in the absence of interleukin 7. J Exp Med 200(9):1179–1187
Ottilie S, Diaz JL, Horne W, Chang J, Wang Y, Wilson G et al (1997) Dimerization properties of human BAD. Identification of a BH-3 domain and analysis of its binding to mutant BCL-2 and BCL-XL proteins. J Biol Chem 272(49):30866–30872
Pahl HL (1999) Activators and target genes of Rel/NF-kappaB transcription factors. Oncogene 18(49):6853–6866
Park MY, Jang HD, Lee SY, Lee KJ, Kim E (2004) Fas-associated factor-1 inhibits nuclear factor-kappaB (NF-kappaB) activity by interfering with nuclear translocation of the RelA (p65) subunit of NF-kappaB. J Biol Chem 279(4):2544–2549
Pineda M, Fernandez E, Torrents D, Estevez R, Lopez C, Camps M et al (1999) Identification of a membrane protein, LAT-2, that Co-expresses with 4F2 heavy chain, an L-type amino acid transport activity with broad specificity for small and large zwitterionic amino acids. J Biol Chem 274(28):19738–19744
Pollard KM, Hultman P (1997) Effects of mercury on the immune system. Met Ions Biol Syst 34:421–440
Pollard KM, Lee DK, Casiano CA, Bluthner M, Johnston MM, Tan EM (1997) The autoimmunity-inducing xenobiotic mercury interacts with the autoantigen fibrillarin and modifies its molecular and antigenic properties. J Immunol 158(7):3521–3528
Pollard KM, Pearson DL, Bluthner M, Tan EM (2000) Proteolytic cleavage of a self-antigen following xenobiotic-induced cell death produces a fragment with novel immunogenic properties. J Immunol 165(4):2263–2270
Price PT, Nelson CM, Clarke SD (2000) Omega-3 polyunsaturated fatty acid regulation of gene expression. Curr Opin Lipidol 11(1):3–7
Quig D (1998) Cysteine metabolism and metal toxicity. Altern Med Rev 3(4):262–270
Rice DC (2008) Overview of modifiers of methylmercury neurotoxicity: chemicals, nutrients, and the social environment. Neurotoxicology 29(5):761–766
Sarmento A, Guilhermino L, Afonso A (2004) Mercury chloride effects on the function and cellular integrity of sea bass (Dicentrarchus labrax) head kidney macrophages. Fish Shellfish Immunol 17(5):489–498
Seil FJ (1998) The extracellular matrix molecule, laminin, induces purkinje cell dendritic spine proliferation in granule cell depleted cerebellar cultures. Brain Res 795(1–2):112–120
Shenker BJ, Guo TL, I O, Shapiro IM (1999) Induction of apoptosis in human T-cells by methyl mercury: temporal relationship between mitochondrial dysfunction and loss of reductive reserve. Toxicol Appl Pharmacol 157(1):23–35
Shenker BJ, Guo TL, Shapiro IM (2000) Mercury-induced apoptosis in human lymphoid cells: evidence that the apoptotic pathway is mercurial species dependent. Environ Res 84(2):89–99
Singh VK, Rivas WH (2004) Prevalence of serum antibodies to caudate nucleus in autistic children. Neurosci Lett 355(1–2):53–56
Stinson CH, Shen DM, Burbacher TM, Mohamed MK, Mottet NK (1989) Kinetics of methyl mercury in blood and brain during chronic exposure in the monkey Macaca fascicularis. Pharmacol Toxicol 65(3):223–230
Takahashi T, Tanaka M, Brannan CI, Jenkins NA, Copeland NG, Suda T et al (1994) Generalized lymphoproliferative disease in mice, caused by a point mutation in the Fas ligand. Cell 76(6):969–976
Tchounwou PB, Ayensu WK, Ninashvili N, Sutton D (2003) Environmental exposure to mercury and its toxicopathologic implications for public health. Environ Toxicol 18(3):149–175
Templeton DM (2000) Metal ions and the cytoskeleton. In: Koropatnick DJ (ed) Molecular biology and toxicology of metals. Taylor and Francis, New York
ten Dijke P, Miyazono K, Heldin CH (2000) Signaling inputs converge on nuclear effectors in TGF-beta signaling. Trends Biochem Sci 25(2):64–70
Tian Y, Green PG, Stamova B, Hertz-Picciotto I, Pessah IN, Hansen R, et al (2009) Correlations of gene expression with blood lead levels in children with autism compared to typically developing controls. Neurotox Res. doi:10.1007/s12640-009-9126-x
Tu Y, Wu S, Shi X, Chen K, Wu C (2003) Migfilin and Mig-2 link focal adhesions to filamin and the actin cytoskeleton and function in cell shape modulation. Cell 113(1):37–47
Warke VG, Nambiar MP, Krishnan S, Tenbrock K, Geller DA, Koritschoner NP et al (2003) Transcriptional activation of the human inducible nitric-oxide synthase promoter by Kruppel-like factor 6. J Biol Chem 278(17):14812–14819
Warren RP, Singh VK, Averett RE, Odell JD, Maciulis A, Burger RA et al (1996) Immunogenetic studies in autism and related disorders. Mol Chem Neuropathol 28(1–3):77–81
Warren RP, Odell JD, Warren WL, Burger RA, Maciulis A, Daniels WW et al (1997) Brief report: immunoglobulin A deficiency in a subset of autistic subjects. J Autism Dev Disord 27(2):187–192
WHO (ed) (1990) Environmental health criteria 101: methylmercury. International Program on Chemical Safety, World Health Organization, Geneva
WHO (ed) (1991) Inorganic mercury. Environmental health criteria 118. International Program on Chemical Safety, World Health Organization, Geneva
Wu Z, Irizarry RA (2004) Preprocessing of oligonucleotide array data. Nat Biotechnol 22(6):656–658 author reply 8
Xu W, Xie Z, Chung DW, Davie EW (1998) A novel human actin-binding protein homologue that binds to platelet glycoprotein Ibalpha. Blood 92(4):1268–1276
Yoon S, Han SS, Rana SV (2008) Molecular markers of heavy metal toxicity—a new paradigm for health risk assessment. J Environ Biol 29(1):1–14
Youle RJ, Strasser A (2008) The BCL-2 protein family: opposing activities that mediate cell death. Nat Rev Mol Cell Biol 9(1):47–59
Zahir F, Rizwi SJ, Haq SK, Khan RH (2005) Low dose mercury toxicity and human health. Environ Toxicol Pharmacol 20:351–360
Zelikoff J, Gardner A (1996) Immunomodulation by metals. In: Chang LW (ed) Toxicology of metals. CRC Press, Boca Raton, pp 783–884
Zhang Y, Derynck R (1999) Regulation of Smad signalling by protein associations and signalling crosstalk. Trends Cell Biol 9(7):274–279
Ziemba SE, McCabe MJ Jr, Rosenspire AJ (2005) Inorganic mercury dissociates preassembled Fas/CD95 receptor oligomers in T lymphocytes. Toxicol Appl Pharmacol 206(3):334–342
Acknowledgments
The authors thank the CHARGE study staff for recruitment, assessment, and data collection and the many families who participated. We also thank the UCD genomics core for processing the samples. This study was supported by NIH grant (I P01 ES11269) (1 R01 ES015359), the U.S. Environmental Protection Agency through the Science to ARCHIVE Results (STAR) program (R829388, R833292), a Cure Autism Now (Autism Speaks) grant, a UC Discovery (Industry-University Cooperative Research Program) grant, and an award from the Clinical and Translational Science Center (CTSC) and MIND Institute at UCD. The authors declare they have no competing financial interests.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below are the links to the electronic supplementary material.
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Stamova, B., Green, P.G., Tian, Y. et al. Correlations Between Gene Expression and Mercury Levels in Blood of Boys With and Without Autism. Neurotox Res 19, 31–48 (2011). https://doi.org/10.1007/s12640-009-9137-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12640-009-9137-7