Abstract
The objective of this study was to examine the correlation between gene expression and lead (Pb) levels in blood in children with autism (AU, n = 37) compared to typically developing controls (TD, n = 15). We postulated that, though lead levels did not differ between the groups, AU children might metabolize lead differently compared to TD children. RNA was isolated from blood and processed on Affymetrix microarrays. Separate analyses of covariance (ANCOVA) corrected for age and gender were performed for TD, AU, and all subjects (AU + TD). To reduce false positives, only genes that overlapped these three ANCOVAs were considered. Thus, 48 probe sets correlated with lead levels in both AU and TD subjects and were significantly different between the groups (p(Diagnosis × log2 Pb) < 0.05). These genes were related mainly to immune and inflammatory processes, including MHC Class II family members and CD74. A large number (n = 791) of probe sets correlated (P ≤ 0.05) with lead levels in TD but not in AU subjects; and many probe sets (n = 162) correlated (P ≤ 0.05) with lead levels in AU but not in TD subjects. Only 30 probe sets correlated (P ≤ 0.05) with lead levels in a similar manner in the AU and TD groups. These data show that AU and TD children display different associations between transcript levels and low levels of lead. We postulate that this may relate to the underlying genetic differences between the two groups, though other explanations cannot be excluded.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Lead (Pb) is a worldwide environmental health hazard that poses a substantial risk to children and adults. The nervous, immune, renal, skeletal, and hematopoetic systems can be affected by lead exposure (Bouton and Pevsner 2000; Landrigan and Todd 1994). The nervous system of children is more sensitive to the toxic effects of lead compared to adults (Needleman 2004). Pediatric lead exposure can affect the development of cognitive, communication, and social functions (Lidsky and Schneider 2003; Gilbert et al. 2005). Some studies have suggested that even low levels of lead can affect children (Needleman 2004; Lanphear 2007; Needleman 2009). Lead, a xenobiotic not known to be a natural component of any biological system, competes with biologically essential metals such as calcium and zinc in metal-binding domains of enzymes, transcription factors, and other proteins (Bouton and Pevsner 2000; Needleman 2004; Yoon et al. 2008). These effects, along with the associated oxidative stress (Adonaylo and Oteiza 1999), likely contribute to lead-induced changes of gene expression in cells (Bouton and Pevsner 2000; Yoon et al. 2008).
Autism (AU) is a developmental disorder characterized by impaired communication, social interactions, and abnormally restricted and repetitive behavior. The etiology of AU may involve a complex combination of environmental, neurological, immunological, and genetic factors (London 2000; Hertz-Picciotto et al. 2006; Ashwood et al. 2006). Though many studies show that lead is neurotoxic, lead has not been implicated in AU. Recent studies suggest that lead exposure when compounded by environmental stress may increase nervous system susceptibility to lead (Virgolini et al. 2005, 2006; White et al. 2007; Cory-Slechta 2005; Cory-Slechta et al. 2004). If true, one might consider AU as a form of “stress” that could alter the metabolism to heavy metal toxicants in children with AU compared to typically developing (TD) children. Previously, using genome wide RNA expression microarrays, we demonstrated that expression (RNA levels) of specific genes in peripheral blood differed between AU and TD controls (Gregg et al. 2008). In this report, we use genome wide RNA expression microarrays to examine the association between gene expression and lead levels in peripheral blood for 2- to 5-year-old AU children compared to TD children.
Methods
Subjects
All of the participating children were recruited as part of the Childhood Autism Risks from Genetics and Environment (CHARGE) study at University of California at Davis. A full description of the study, including recruitment and the assessment criteria, has been published (Hertz-Picciotto et al. 2006). AU diagnoses were assessed using the Autism Diagnostic Interview-Revised (ADI-R) (Lord et al. 1994, 1997), and the Autism Diagnostic Observation Schedules (ADOS, Lord et al. 2000, 2003). The final diagnosis of AU was defined as meeting criteria on the communication, social, and repetitive behavior domains of the ADI-R, and scoring at or above the total cutoff for autistic disorder on the ADOS module 1 or 2. The TD controls were randomly sampled from California state birth certificate files. The Social Communication Questionnaire, Mullen Scales of Early Learning and Vineland Adaptive Behavioral Scales, along with a developmental pediatrician interview and examination were used to exclude putative TD subjects with behavioral, developmental or autism spectrum disorder (ASD) abnormalities.
There were 37 children with AU and 15 TD children from the general population in this study. The average age of AU subjects was 44.2 ± 10 months and that for TD was 41.2 ± 6 months, which was not significantly different (t-test, P > 0.05). There were 32 males and 5 females in the AU group of whom 17 were Hispanic, 16 were white and 4 were other races. There were 11 males and 4 females in the TD group of whom 8 were Hispanic, 5 were Caucasian, and 2 were other races. The male predominance reflects the prevalence of the AU in males being higher than females (Hertz-Picciotto et al. 2006; Gillberg and Wing 1999) and the CHARGE study matching of population controls to the projected sex ratio of cases. We included both male and female subjects and corrected gender effects in the analyses.
Blood Lead Measurement
Blood samples for lead were collected in ethylenediaminetetraacetic acid (EDTA) vacutainer tubes. A portion of the blood (0.5 ml) designated for metal analysis was then stored at −20°C. Lead was measured using an Agilent 7500i (Palo Alto, CA) Inductively Coupled Plasma Mass Spectrometer (ICP-MS) in the Department of Civil and Environmental Engineering, UC Davis. Detailed methods were included in a previous publication (Hertz-Picciotto et al. 2009). Samples were thawed and diluted 20-fold with water containing 1% nitric acid (trace metal grade, Fisher Scientific, Pittsburgh, PA), 0.05% Triton x-100 (Fisher), and 100 ppb Tb (Spex Certiprep, Methuen, NJ) as an internal standard. Samples were loaded at 0.4 ml/min using a peri-staltic pump with 1.2 l/min Argon (Ar) carrier gas through a Babbington-style nebulizer into a Peltier-cooled double-pass spray-chamber at 2°C. Then, 1 l/min auxiliary Ar and 12 l/min plasma gas Ar were added for a total of 14 l/min and separated from Ni cones by a sampling depth of 8.5 mm. Validation used the Performance Testing standards from New York State (Wadsworth Center). The limit of detection (LOD) for lead was 0.006 μg/dl.
RNA Isolation and Microarray Processing
RNA isolation and microarray processing were performed as described previously (Gregg et al. 2008). Briefly, 15 ml blood was collected into six PAXgene vacutainer tubes (Qiagen; Valencia, CA). Total RNA was isolated using the PAXgene blood RNA kit (Qiagen; Valencia, CA) according to the manufacturer’s instructions. RNA quality and purity were analyzed by spectrophotometry using the Nanodrop ND-1000, and RNA integrity was analyzed using the Agilent 2100 Bioanalyzer. Samples achieved A260/A280 absorbance ratios of purified RNA exceeding 2.0, the 28S/18S rRNA ratios were equal to or exceeded 1.5, and the RNA Intergrated Number (RIN) exceeded 8.0. Gene expression was assessed on the Human U133 Plus 2.0 GeneChip microarrays (Affymetrix; Santa Clara, CA) according to the standard Affymetrix expression analysis protocols. The microarrays detected over 54,000 probe sets which assessed over 38,500 human genes (Affymetrix manual). In our study, we use the term “probe sets” for a designated group of oligonucleotides on a microarray, and the term “genes” for known, annotated genes.
Statistical Analyses
The analyses were performed using Partek Genomics Suite, version 6.4, release 6.09.0422 (Partek Inc., St. Louis, MO, USA). Gene expression data were normalized using GC-RMA. Analysis of covariance (ANCOVA) was performed and the partial correlations were estimated between gene expression and log2-transformed lead levels, while controlling the effects of age and gender. The partial correlation is the correlation that remains between the dependent variable (gene expression) and the independent variable of interest (log2 Pb) after removing the correlation that is due to their mutual association with the other independent variables (age and gender). We performed an ANCOVA on the three models shown in Fig. 1. The variables for these three models are:
-
Model 1 (TD samples only): log2 Pb (continuous variable), Age (continuous variable), Gender (categorical variable).
-
Model 2 (AU samples only): log2 Pb (continuous variable), Age (continuous variable), Gender (categorical variable).
-
Mode 3 (AU and TD samples combined): log2 Pb (continuous variable), Age (continuous variable), Gender (categorical variable), Diagnosis (categorical variable), Diagnosis × log2 Pb (interaction).
Venn diagrams showing the work flow for the statistical analyses. GE gene expression; TD typically developing children from the general population; AU Children with autism. Model 1 identified genes whose expression significantly correlated (p(log2 Pb) < 0.05) with lead levels in TD after accounting for partial correlations for age and gender. Model 2 identified genes whose expression significantly correlated (p(log2 Pb) < 0.05) with lead levels in AU after accounting for partial correlations for age and gender. Model 3 identified genes whose expression significantly differently correlated with lead levels for AU compared to TD based on the interaction of diagnosis with lead level after adjustment for age and gender. Gene list A represents genes whose expression correlates significantly (p(log2 Pb) ≤ 0.05) with log2 Pb levels in each of AU and TD models separately, and the partial correlation coefficients are significantly different from each other in combined model(p(Diagnosis × log2 Pb) ≤ 0.05). Gene list B represents genes whose expression correlates (p(log2 Pb) ≤ 0.05) with log2 Pb levels in each of AU and TD models separately, but the partial correlation coefficients are not significantly different from each other in combined model. Gene list C represents genes whose expression significantly correlates with log2 Pb levels in TD (p(log2 Pb) ≤ 0.05) but not in AU subjects, and the partial correlation coefficients are significantly different from each other in combined model (p(Diagnosis × log2 Pb) ≤ 0.05). Gene list D represents genes whose expression significantly correlates with log2 Pb levels in AU (p(log2 Pb) ≤ 0.05) but not in TD subjects, and the partial correlation coefficients are significantly different from each other in combined model (p(Diagnosis × log2 Pb) ≤ 0.05)
In order to decrease the numbers of false positives, we reasoned that the genes that demonstrated overlaps between the ANCOVA Models would be the most reliable. These gene overlaps are shown in Fig. 1 (ListA-D) and represent the following:
-
Gene list A represents genes whose expression correlated with lead levels in each of AU and TD models separately (p(log2 Pb) ≤ 0.05), and the partial correlation coefficients were statistically different between the two groups in combined model (p(Diagnosis × log2 Pb) ≤ 0.05).
-
Gene list B represents genes whose expression correlated with lead levels both in AU and TD models separately (p(log2 Pb) ≤ 0.05) without any significant difference in the partial correlation coefficients between the two groups in combined model (p(Diagnosis × log2 Pb) > 0.05).
-
Gene list C represents genes whose expression significantly correlates with lead levels in TD (p(log2 Pb) ≤ 0.05) but not in AU models, and the partial correlation coefficients were statistically different between the two groups in combined model (p(Diagnosis × log2 Pb) ≤ 0.05).
-
Gene list D represents genes whose expression significantly correlates with lead levels in AU (p(log2 Pb) ≤ 0.05) but not in TD models, and the partial correlation coefficients were statistically different between the two groups in combined model (p(Diagnosis × log2 Pb) ≤ 0.05).
As described above, each of these gene lists was adjusted to account for partial correlations due to age and gender. Gene list A + C + D contained genes, whose expression differently correlated with blood lead levels, only Gene list B contained genes, whose expression correlated with lead levels in a similar manner.
Assessment of Biological Significance of Identified Genes
The network, molecular functions, and toxicant functions were analyzed using Ingenuity Pathways Analysis (IPA, Ingenuity® Systems, www.ingenuity.com). The datasets (gene lists) were uploaded into IPA. Each identifier was mapped to its corresponding gene object and overlaid onto a global molecular network developed from information contained in the Ingenuity knowledge base. IPA network analyses answer the question of what relationships exist between the genes in the dataset. The networks were ranked by a score according to how relevant they are to the genes in the input dataset. The score takes into account the number of focus genes in the network and the size of the network to approximate how relevant this network is to the original list of focus genes. The greater the number of network eligible molecules in a network, the higher the score (lower the P value) will be. The molecular and cellular functions analysis identified the molecular and cellular functions that were most significant to the dataset. The toxicant functions analysis identified the xenobiotic pathways that were most significant to the dataset. Fisher’s exact test was used to calculate a P-value to determine the probability that the association between the genes in the pathways and the genes in each list is explained by chance alone. A P ≤ 0.05 was considered to be statistically significant for overrepresentation of the molecules in a given process.
Results
Blood Lead Levels in AU and TD
There were no significant differences in blood lead levels between AU and TD (t-test, P = 0.97). The mean blood lead level for AU was 1.30 ± 1.01 μg/dl (Mean ± SD), and for TD was 1.30 ± 0.58 μg/dl (Mean ± SD). The geometric mean blood lead level for AU was 1.03 μg/dl, and for TD was 1.16 μg/dl. Blood lead levels ranged from 0.32 to 5.21 μg/dl in AU subjects, and ranged from 0.37 to 2.99 μg/dl in TD subjects. No blood lead level was higher than 10 μg/dl in any subject. A log2-transformation was applied to blood lead levels due to the skewed distribution values in both AU and TD (Supplementary Fig. 1). The distribution of the log2-transformed lead levels followed a normal distribution (Kolmogorov–Smirnov Test P = 0.71, 0.92, and 0.89 for TD, AU and TD + AU, respectively).This resulted in a more linear relationship between gene expression and blood lead levels.
Genes Whose Expression Correlated with Lead Levels
Figure 2 shows the total numbers of probe sets derived from the ANCOVAs shown in Fig. 1 and shows how many genes overlapped for each analysis. The ANCOVA analysis for TD showed that 1,712 probe sets significantly (p(log2 Pb) ≤ 0.05) correlated with log2 Pb levels (Fig. 2, bottom left circle). The ANCOVA analysis for AU showed 1,829 probe sets that significantly (p(log2 Pb) ≤ 0.05) correlated with log2 Pb levels (Fig. 2, bottom right circle). The ANCOVA analysis for AU and TD combined group showed that expression of 2,172 probe sets differed significantly between the AU and TD groups as a function of log2 Pb levels (p(diagnosis × log2 Pb) < 0.05; Fig. 2, top circle).
Venn diagrams showing the numbers of probe sets obtained for each of the analyses performed in Fig. 1. List A: Only 48 probe sets were shared between the three ANCOVAs. These genes were significantly correlated (p(log2 Pb) ≤ 0.05) with lead levels in both AU and TD groups, and were significantly different from each other (p(Diagnosis × log2 Pb) ≤ 0.05). List B: A total of 30 probe sets were significantly correlated (P ≤ 0.05) with lead levels in both the AU and TD groups, and were not significantly different from each other. List C: A total of 791 probe sets significantly correlated (P ≤ 0.05) with lead levels in TD (P ≤ 0.05) (Supplementary Table 4) but not in AU. List D: A total of 162 probe sets significantly correlated (P ≤ 0.05) with lead levels in AU (P ≤ 0.05) (Supplementary Table 5) but not in TD
There were four gene lists that represented the different overlaps for the ANCOVAs which are designated A, B, C, and D in Fig. 1. The numbers of genes in each overlap area are shown in Fig. 2.
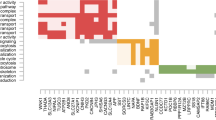
Gene list A (Fig. 1) contained 48 probe sets (Fig. 2) that overlapped the three ANCOVA models. The expression of these genes significantly correlated with log2 Pb levels in both the AU and TD groups (p(log2 Pb) ≤ 0.05 for both groups)—but the partial correlation coefficients were significantly different from each other (p(diagnosis × log2 Pb) < 0.05) (Supplementary Table 1). The 48 probe sets represent 31 annotated genes and the partial correlations for these genes are shown for the AU and TD groups in Fig. 3. A number of human leukocyte antigen (HLA) genes, including HLA-DRB subtype and the major histocompatibility complex (MHC) class II-associated invariant chain CD74 are shown in the heat map. ANXA2 and ANXA4, two calcium-dependent phospholipid binding genes, are also shown (Fig. 3). Note that most of the genes are negatively (blue) correlated with blood lead levels in the TD subjects but positively (red) correlated with blood lead levels in the AU subjects (Fig. 3). Only two genes, TUBGCP3 and SH3BP2, are positively correlated with blood lead levels in TD and negatively correlated in AU. The partial correlations for three genes, ALDH3A2 and PCDHB11 and AMN, were negatively correlated with log2 Pb levels in TD and negatively correlated with log2 Pb levels in AU, but were significantly different (p(diagnosis × log2 Pb) ≤ 0.05) between the groups. The partial correlations in Fig. 3 ranged from −0.72 to +0.54 for TD and from −0.47 to +0.53 for AU. The microRNA targets for these 48 genes are listed in Supplementary Table 2.
A heat map of the partial correlations of the 31 annotated genes that were represented by 48 probe sets identified in gene List A in Fig. 2. Genes are shown in the rows, and diagnostic classes (AU and TD) are in columns. Negative correlations are shown in blue and positive correlations are shown in red (see calibration bar at bottom). These genes represent those that significantly correlated with lead levels in both AU and TD subjects (p(log2 Pb) ≤ 0.05 for each group). In addition, the partial correlation coefficients for these genes were statistically significantly different between the TD and AU subjects when the interaction of diagnosis and lead levels was examined (p(Diagnosis × log2 Pb) < 0.05). HLA-DRB* is the abbreviation for HLA-DRB1/2/3/4/5. IGH* is the abbreviation for IGH@/A1/A2/G1/G3/M/V. (Color figure online)
Gene list B (Fig. 1) contained 30 probe sets (Fig. 2) that were shared between the TD and AU ANCOVAs. These 30 probe sets represent those that were correlated with log2 Pb levels in both AU and TD subjects, and were not statistically different (p(diagnosis × log2 Pb) > 0.05). These genes included the Aryl hydrocarbon receptor nuclear translocator (ARNT), an ATPase family member (ATAD), polyserase 3(POL3S), peroxiredoxin 2(PRDX2), dynein (axonemal, light chain 1) (DNAL1), secretory carrier membrane protein 1 (SCAMP1) and alpha synuclein (SNCA) (Supplementary Table 3).
Gene list C (Fig. 1) had 791 probe sets (Fig. 2) that were significantly correlated (P ≤ 0.05) with blood lead levels in the TD subjects but not in the AU subjects (Supplementary Table 5). This is shown in the cluster in Fig. 4. Of these 791 probe sets, a large number of probe sets (250) are positively correlated (red) with blood lead levels and an even larger number of probe sets (541) are negatively correlated (blue) with blood lead levels in the TD subjects. However, these probe sets showed no correlation with blood lead levels in the AU subjects (Fig. 4). The glutathione S-transferase gene (GSTT1) is in this list (Supplementary Table 4) and negatively correlated with blood lead levels in TD subjects (Partial correlation: −0.62).
A heat map showing the partial correlations of the 791 probe sets identified in gene List C in Fig. 2. Probe sets are in rows and the AU and TD diagnostic groups are in the columns. Negative correlations are shown in blue and positive correlations are shown in red (see calibration bar at bottom). These 791 probe sets represent genes that significantly correlated (p(log2 Pb) ≤ 0.05) with lead levels in TD and not in AU subjects. Note high positive and negative correlations in the TD group and lack of correlations in the AU group. (Color figure online)
Gene list D (Fig. 1) had 162 probe sets (Fig. 2) that were significantly correlated (P ≤ 0.05) with blood lead levels in the AU subjects but not in the TD subjects (Supplementary Tables 5). This is shown graphically in the cluster in Fig. 5. Of these 162 probe sets, there are a number of probe sets (76) that are positively correlated (red) with lead levels in AU subjects; and a similar number of probe sets (86) that are negatively correlated (blue) with blood lead in the AU subjects. The partial correlations in most of genes in the AU (Fig. 5, right column) subjects tended to be inverse in sign to the partial correlations in the TD subjects (Fig. 5, left column), though these were not statistically significant (p(log2 Pb) > 0.05).
A heat map showing the partial correlations of the 162 probe sets identified in List D in Fig. 2. Probe sets are shown in the rows and the AU and TD groups in the columns. Negative correlations are shown in blue and positive correlations are shown in red (see calibration bar at bottom). These 162 probe sets represent those that significantly correlated (p(log2 Pb) ≤ 0.05) with lead levels in children with AU and not in TD subjects. Note high positive and negative correlations in the AU group and modest inverse correlations in the TD group. (Color figure online)
Biological Significance
Network Functions of the Genes Whose Expression Correlated Differently with Lead Levels in AU and TD (List A)
An IPA network function analysis was performed for the 48 probe sets that correlated with blood lead levels in the AU and TD groups, but were significantly different between the groups. Figure 6 illustrates the top network which includes many genes involved in immunological and inflammatory disease processes. Most of the genes in this network showed a positive correlation with log2 Pb levels in the AU subjects and a negative correlation with log2 Pb levels in TD subjects. Only ALDH3A2 showed a negative correlation in AU and positive correlation in TD.
The top network that came from the 48 probe sets that correlated significantly (p(log2 Pb) ≤ 0.05) with lead levels in both AU and TD, but for which the correlation coefficients were significantly different between the two groups (p(Diagnosis × log2 Pb) < 0.05). This network is associated with immunological and inflammatory disease processes. AU (+) (Red) represents probe sets that positively correlated with lead levels in AU. AU (–) (Blue) represent probe sets that negatively correlated with lead levels in AU. (Color figure online)
Molecular and Cellular Function Analysis for All of the Genes Whose Expression Correlated with Blood Lead Levels Differently in AU and TD (Lists A, C, D)
Given that the genes that correlated with lead levels in AU and TD were so different, it was of interest to know what molecular functions and pathways are differentially affected. Thus, the three gene lists that represented the differentially correlated genes in AU compared to TD (Fig. 2; Lists A, C, D) were used as inputs for IPA analysis. We did not include list B because these are the genes that correlate with lead levels in a similar fashion in AU and TD subjects.
Table 1 lists the top five overrepresented molecular and cellular functions for genes whose expression correlated with blood lead levels differently in AU and TD (Lists A, C, D). For genes that correlated with lead levels differently in AU and TD (List A) many participate in cell–cell signaling, antigen presentation, cell cycle, development and growth, and proliferation (Table 1). The genes that correlated with lead levels in TD (List C) were associated with post-translational modification, protein synthesis, cell morphology, cell-to-cell signaling and cellular assembly and organization (Table 1). The genes that correlated with lead levels in AU subjects (List D) were associated with cellular compromise, drug metabolism, lipid metabolism, small molecule biochemistry, and DNA replication, recombination and repair (Table 1).
Xenobiotic Pathway Analysis for All Genes Whose Expression Correlated with Blood Lead Levels Differently in AU and TD (Lists A, C, D)
Figure 7 shows mitochondrial dysfunction pathways (P = 0.014) were overrepresented in the 48 probe sets that correlated with lead in AU and TD but were different between the groups (List A). Fatty acid metabolism, PXR/RXR activation, G2/M transition of the cell cycle pathways, TR/RXR action, and LPS/IL-1meditated inhibition of RXR function were overrepresented in the 791 probe sets that correlated with lead levels in TD subjects (ranging from P = 0.039 to P = 0.00001) (List C). There was no xenobiotic pathway significantly overrepresented in the 162 probes that correlated with lead levels in AU (mitochondrial dysfunction, P = 0.17) (List D).
Xenobiotic pathway analyses for genes whose expression correlated with lead levels differently in AU compared to TD (List A, C, D). (I). Mitochondrial dysfunction pathways (MD) were overrepresented for gene list A. These 48 probe sets represented genes whose expression correlated with lead levels in both AU and TS groups, but the partial correlation coefficients were statistically different from each other. (II) Pathways that were overrepresented in gene list C that had 791 probe sets whose expression correlated with lead levels in TD, but not AU. LPS/IL1: lipopolysaccharide and interleukin pathways. TR/RXR: thyroid hormone receptor (TR)/retinoic acid receptor (RXR) activation. G2/M: cell cycle transition and cell cycle pathways. PXR/RXR: pregnane X receptor (PXR)/retinoic acid receptor activation. FAM: fatty acid metabolism pathways. There were no statistically significant xenobiotic pathways detected in children with AU. (Color figure online)
Discussion
One of the most significant findings of this study is that the expression of many genes correlated with circulating blood lead concentrations even at low lead levels in AU and TD children (List A, B, C, D). The finding that there are correlations between expression of hundreds of genes in blood with blood lead levels in both groups of children supports the notion that increasing levels of lead, even at low levels, are associated with differences of transcription in blood. Thus, even low levels of lead may produce biological effects in humans. Whether these are toxicological responses or adaptive/protective responses may depend upon the environmental context and genetic context in which they occur.
The other significant finding of the study is that almost all of the genes that correlated with lead levels in TD subjects did not correlate in AU subjects. Moreover, almost all of the genes that correlated with lead levels in AU subjects did not correlate in TD subjects. Of the thousand genes that correlated with lead in this study, only a very few (n = 78, list A + list B) overlap between AU and controls. Even among these few genes that correlate with lead levels and are regulated in AU and controls, the majority of the partial correlation coefficients (n = 48, list A) are statistically different from one another. Thus, the data show that the association of gene transcription with lead levels differs in children with AU compared to controls.
A major question, therefore, is whether this has any relationship to symptoms of AU. The present data cannot provide the answer. Lead levels have been declining steadily in the environment since the late 1970s when lead additives to gasoline were phased out, whereas the incidence of AU has been rising since then (Hertz-Picciotto and Delwiche 2009). Thus, lead probably does not account for the increasing incidence of AU. In our another study, the correlation of gene expression with mercury levels in children with AU is also very different from matched controls (Stamova et al. submitted). Thus, the associations between transcription and levels for both heavy metals differ in AU compared to TD, suggesting there is not a specific abnormality associated with a given heavy metal. Though it is difficult to ascribe cause or effect, individuals with AU may have different genetic backgrounds from controls, and that when presented with a heavy metal like lead have different transcriptional programs. If this is the case, then the differing transcriptional programs following exposure to lead (and mercury) in children with AU and TD controls are likely to be adaptive in order to successfully deal with the heavy metal. Other explanations, however, cannot be ruled out: differences in exposures, epigenetic marks or risk factors like parental age. Though it seems unlikely that the findings of this study directly relate to the symptoms of AU, it is unknown whether these different transcriptional programs might influence the phenotypic variation in AU or even in TD controls (Tang et al. 2004).
Blood Lead Levels
There were no significant differences in blood lead levels between children with AU and controls in this study. The geometric mean blood lead level for AU was 1.03 μg/dl, and for TD was 1.16 μg/dl. These are a little below the national average of 1.9 μg/dl (geometric mean) for 1- to 5-year old children in the United States in 1999–2004 (Jones et al. 2009). No blood lead level was above the lead toxicity threshold of 10 μg/dl established by the United States Centers for Disease Control and Prevention (CDC). Thus, any correlations between gene expression and lead levels are not due to high levels of lead in this study. Although blood lead levels in children have been declining in the United States since late 1970s, some authors have suggested that there is no “safe level” of lead and that neurobehavioral deficits and learning impairments occur even when blood lead levels are below 10 μg/dl (Lanphear 2007; Needleman 2009; Needleman 2004; Needleman and Landrigan 2004; Rossi 2008; Lanphear et al. 2005). Our data provide evidence for a correlation between gene expression and blood lead levels below 10 μg/dl in blood of humans.
Functions of Genes Whose Expression Correlated with Lead Levels Differently in AU and TD (List A)
Of the 48 probe sets that significantly correlated with log2 Pb levels in both AU and TD groups, but the partial correlation coefficients were statistically different, many were associated with immunological and inflammatory processes (Fig. 3). In the CNS, lead treated rat astrocytes express a number of pro-inflammatory cytokines including IL-1β, IL-6, and TNFα (Struzynska et al. 2007). In human peripheral mononuclear cells, lead regulates TNF-alpha and TNF-alpha receptor expression (Guo et al. 1996). For the top network identified for the 48 probe sets differentially regulated between AU and TD, TNF was central to the pathway—though TNF RNA levels themselves were unchanged. Notably, lead is reported to produce immunomodulatory effects by increasing MHC molecule surface expression in both murine (Ia) and human HLA antigen presenting cells (Guo et al. 1996; McCabe and Lawrence 1991). Thus, the correlations of HLA subtype gene levels with lead levels in our study support these previous findings.
Dysregulation of the immune response has also been associated with AU. Abnormal cytokine levels, immunoglobulin levels, complement levels, and decreased lymphocyte numbers have been described in children with AU (Ashwood et al. 2006; Warren et al. 1997; Warren et al. 1996; Gupta et al. 1996; Singh and Rivas 2004; McCabe et al. 1991; Corbett et al. 2007). In addition, HLA-DRB1 and complement C4 alleles are also associated with AU (Ashwood et al. 2006; Warren et al. 1997; Warren et al. 1996; Gupta et al. 1996). The HLA system plays a central role in immune function. In humans, peptide antigens are presented to T cells in the context of HLA molecules (Nepom and Erlich 1991). We found several HLA genes that correlated with lead levels differently in AU compared to TD subjects. These included HLA-DRB subtypes and the MHC class II-associated invariant chain CD74 which is associated with intracellular sorting of MHC class II molecules (Fig. 3). In addition, the antigen presentation function was overrepresented in gene list A (Table 1). These immune genes and pathways that correlate differently with lead levels in AU compared to TD controls could indicate: (1) the immune aberration observed in AU might be due in part to lead or another toxicant; (2) different immune responses in AU compared to TD controls may alter the transcriptional association to various environmental factors including lead in these groups; (3) lead or other toxicants may interact with the underlying genetic background in AU and predispose to immune dysfunction.
Lead competes with calcium and zinc for binding sites on proteins, and thus affects a variety of cellular pathways and downstream gene expression (Bouton and Pevsner 2000). In our study, two annexin protein members, ANXA2 and ANXA4, correlated with lead levels in both AU and TD, and the partial correlations were significantly different in AU and TD. Annexins are a family of calcium-dependent phospholipid binding proteins which are involved in signal transduction and other cellular processes. Expression of ANXA1 and ANXA5 increase in lead-exposed astrocytes, and ANXA5 can be directly bound and activated by nanomolar concentrations of lead (Bouton et al. 2001). Since all members of the annexin protein family have similar functional and structural domains as those in ANXA1 and ANXA5, lead may also competitively bind ANXA2 and ANXA4 and influence signal transduction. Of interest in AU, ANXA4 and ANXA5 are important in the maintenance of pregnancy by preventing activation of blood coagulation. Recent data suggest that ANXA4 enters the blood just after pregnancy probably to prevent coagulation, and ANXA5 is expressed in the placenta probably to prevent coagulation (Masuda et al. 2004). ANXA4 inhibits Staphylococcus aureus attachment to human macrophages and may play a role in other infections (Gotoh et al. 2005).
The xenobiotic pathway analyses showed that mitochondrial dysfunction was overrepresented for genes whose expression correlated with lead levels differently in AU and TD (List A, Supplementary Table 1). Mitochondria play a central role in energy metabolism and other functions including cell death, cell proliferation, steroid synthesis, heme synthesis and calcium signaling (McBride et al. 2006; Green 1998; Rossier 2006). Lead exposure alters the mitochondrial structure in tissues taken from lead-intoxicated animals and in human biopsy specimens (Goyer 1968; Goyer et al. 1968). Competing with calcium, lead is taken up into mitochondria in an energy dependent manner (Walton 1973; Walton and Buckley 1977; Kapoor et al. 1985). Future studies are needed to determine if the differences in expression of the mitochondrial genes that correlate with lead in AU compared to control children translate into dys-regulated mitochondrial function in AU.
Functions of Genes Whose Expression Correlated with Blood Lead Levels in AU or TD (Lists C and D)
Our data also show that several xenobiotic pathways are overrepresented in genes correlating only with lead levels in TD controls, including fatty acid metabolism, PXR/RXR activation, G2/M transition of the cell cycle pathways, TR/RXR action, LPS/IL-1meditated inhibition of RXR function (Fig. 7). Of interest, there were no statistically significant toxicant pathways detected in children with AU (Fig. 7), as opposed to TD children who had many such xenobiotic pathways.
Fatty acids are an integral part of cell membranes and are an energy reserve for cells. Lead can alter fatty acid composition and lipid peroxidation in animal liver (Knowles and Donaldson 1996; Lawton and Donaldson 1991) and fatty acids are implicated in neurodevelopmental disorders including AU (Richardson and Ross 2000; Wiest et al. 2009). In earlier work from the CHARGE study, we found that plasma fatty acid profiles in children with AU differed from controls (Wiest et al. 2009). This study shows that fatty acid metabolism genes correlated with lead levels in TD and not in AU which may provide additional support for fatty acid metabolism abnormalities in AU. Notably, 12 molecules correlated with lipid metabolism in AU but not TD subjects (Table 1), further suggesting dysfunction of lipid and fatty metabolism in AU compared to TD subjects.
GSTT1 expression, which correlated with lead levels in TD but not AU subjects (Supplementary Table 4), is involved in LPS/IL-1 meditated inhibition of RXR function pathway in our study. GSTs are a family of enzymes that catalyze the conjugation of reduced glutathione to a variety of electrophilic and hydrophobic compounds (Wright et al. 1998). GSTs are major mediators of cellular responses to toxicants and stress (Bouton and Pevsner 2000; Qu et al. 2002). The expression of a number of GST isoforms changes in response to both acute and chronic lead exposure (Daggett et al. 1998; Oberley et al. 1995; Moser et al. 1995). GST-P is induced in rat liver and kidney by lead, and the activation of the GST-P gene is mediated in part by TDEI and AP-1 (Suzuki et al. 1996). In the kidney, the increase in GST by lead precedes cellular damage, and thus may serve as a biomarker of lead exposure (Wright et al. 1998). GSTT1 was also significantly increased in metallothionein (MT)-null mice after lead exposure (Qu et al. 2002). Though GSTT1 expression correlates with lead levels in TD but not AU subjects in this study, the significance of this is uncertain.
Genes that Correlate Similarly with Lead Levels in Both AU and TD (List B)
Prior to this study, one might well have hypothesized that the transcriptome that correlated with lead levels in AU and TD subjects would be the same. Thus, finding only 30 transcripts correlate similarly with lead levels in both AU and TD subjects does not support this hypothesis. However, the genes that do correlate with lead in both groups are of interest. Dynein, which correlated with lead levels in both groups, is one of the microtubule-based motors (Rubinsztein et al. 2005) and might be affected by heavy metals—though this does not appear to have been studied. Of more interest, alpha synuclein expression negatively correlates with lead levels in both TD subjects (Partial correlation = −0.62) and in AU subjects (Partial correlation = −0.32). This protein has been implicated in the degenerative synucleinopathies of brain including Parkinsons disease and Alzheimer’s disease (Eller and Williams 2009; Jellinger 2009; Crews et al. 2009), and heavy metals like lead have been sometimes been implicated in these conditions (White et al. 2007).
Limitations of the Study
It is important to emphasize that the current data must be interpreted with caution. Though the age, gender, and race were similarly matched, our group sizes were small with unequal numbers of subjects in each group. This could underlie some differences between the groups. Since the data are cross-sectional, it is impossible to draw strong conclusions about any of the associations observed unless increases and decreases of lead levels over time were shown to be correlated with increases and decreases of gene expression over time. Since this is a one-time study, the correlation of an individual gene might not be replicated in future studies due to stochasticity.
Of equal concern, other differences between cases and controls are possible, and adjustment for those differences would alter the findings. We have reported differences in lipid metabolism, sleep patterns, and pesticide exposures of cases versus controls in the CHARGE Study (Wiest et al. 2009; Krakowiak et al. 2008). These differences may be confounding the relationships of gene expression to lead that might differ in AU and TD groups.
Another limitation of the study is that the circulating lead represents a small fraction of the body burden. The largest store of lead is in bone where it turns over slowly. Thus, the transcriptional associations with lead levels in blood probably do not reflect those in the bone, and probably do not reflect those in brain and other organs.
Finally, an unresolved issue was how best to correct for multiple comparisons. The Benjamini-Hochberg False Discovery (FDR) rate approach is most commonly used for such corrections in microarray studies because of the very large number of variables. However, the FDR has shortcomings since it assumes all genes are independently expressed, which is not the case (Zhang 2006). Thus, we chose to use two criteria to decrease the numbers of false positive genes: partial correlations had to have a P ≤ 0.05 level using an ANCOVA; and the identified significant genes had to overlap for at least two of the ANCOVAs performed. However, if one simply ignores any correction for multiple comparisons, then 1,712 genes correlated with lead in TD and 1,829 genes correlated with lead in AU, but only 78 genes were shared between the two analyses. By not correcting for multiple comparisons, this should maximize the genes that overlapped between AU and TD which in this case is 78 out of >1,000 genes. Thus, the major conclusion of the study is unchanged whether a correction for multiple comparisons is applied or not. The conclusion is: almost all of the genes whose expression correlates with lead levels in children with AU differ from those that correlate with lead levels in TD children.
References
Adonaylo VN, Oteiza PI (1999) Lead intoxication: antioxidant defenses and oxidative damage in rat brain. Toxicology 135(2–3):77–85
Ashwood P, Wills S, Van de Water J (2006) The immune response in autism: a new frontier for autism research. J Leukoc Biol 80(1):1–15
Bouton CM, Pevsner J (2000) Effects of lead on gene expression. Neurotoxicology 21(6):1045–1055
Bouton CM, Hossain MA, Frelin LP, Laterra J, Pevsner J (2001) Microarray analysis of differential gene expression in lead-exposed astrocytes. Toxicol Appl Pharmacol 176(1):34–53
Corbett BA, Kantor AB, Schulman H, Walker WL, Lit L, Ashwood P et al (2007) A proteomic study of serum from children with autism showing differential expression of apolipoproteins and complement proteins. Mol Psychiatry 12(3):292–306
Cory-Slechta DA (2005) Studying toxicants as single chemicals: does this strategy adequately identify neurotoxic risk? Neurotoxicology 26(4):491–510
Cory-Slechta DA, Virgolini MB, Thiruchelvam M, Weston DD, Bauter MR (2004) Maternal stress modulates the effects of developmental lead exposure. Environ Health Perspect 112(6):717–730
Crews L, Tsigelny I, Hashimoto M, Masliah E (2009) Role of synucleins in Alzheimer’s disease. Neurotox Res 16(3):306–317
Daggett DA, Oberley TD, Nelson SA, Wright LS, Kornguth SE, Siegel FL (1998) Effects of lead on rat kidney and liver: GST expression and oxidative stress. Toxicology 128(3):191–206
Eller M, Williams DR (2009) Biological fluid biomarkers in neurodegenerative parkinsonism. Nat Rev Neurol 5:561–570
Gilbert ME, Kelly ME, Samsam TE, Goodman JH (2005) Chronic developmental lead exposure reduces neurogenesis in adult rat hippocampus but does not impair spatial learning. Toxicol Sci 86(2):365–374
Gillberg C, Wing L (1999) Autism: not an extremely rare disorder. Acta Psychiatr Scand 99(6):399–406
Gotoh M, Takamoto Y, Kurosaka K, Masuda J, Ida M, Satoh A et al (2005) Annexins I and IV inhibit Staphylococcus aureus attachment to human macrophages. Immunol Lett 98(2):297–302
Goyer RA (1968) The renal tubule in lead poisoning. I. mMitochondrial swelling and aminoacidura. Lab Invest 19(1):71–77
Goyer RA, Krall A, Kimball JP (1968) The renal tubule in lead poisoning. II. In vitro studies of mitochondrial structure and function. Lab Invest 19(1):78–83
Green DR (1998) Apoptotic pathways: the roads to ruin. Cell 94(6):695–698
Gregg JP, Lit L, Baron CA, Hertz-Picciotto I, Walker W, Davis RA et al (2008) Gene expression changes in children with autism. Genomics 91(1):22–29
Guo TL, Mudzinski SP, Lawrence DA (1996a) The heavy metal lead modulates the expression of both TNF-alpha and TNF-alpha receptors in lipopolysaccharide-activated human peripheral blood mononuclear cells. J Leukoc Biol 59(6):932–939
Guo TL, Mudzinski SP, Lawrence DA (1996b) Regulation of HLA-DR and invariant chain expression by human peripheral blood mononuclear cells with lead, interferon-gamma, or interleukin-4. Cell Immunol 171(1):1–9
Gupta S, Aggarwal S, Heads C (1996) Dysregulated immune system in children with autism: beneficial effects of intravenous immune globulin on autistic characteristics. J Autism Dev Disord 26(4):439–452
Hertz-Picciotto I, Delwiche L (2009) The rise in autism and the role of age at diagnosis. Epidemiology 20(1):84–90
Hertz-Picciotto I, Croen LA, Hansen R, Jones CR, van de Water J, Pessah IN (2006) The CHARGE study: an epidemiologic investigation of genetic and environmental factors contributing to autism. Environ Health Perspect 114(7):1119–1125
Hertz-Picciotto I, Green PG, Delwiche L, Hansen R, Walker C, Pessah IN (2009) Blood mercury concentrations in CHARGE study children with and without autism. Environ Health Perspect. doi:10.1289/ehp.0900736
Jellinger KA (2009) Recent advances in our understanding of neurodegeneration. J Neural Transm 116(9):1111–1162
Jones RL, Homa DM, Meyer PA, Brody DJ, Caldwell KL, Pirkle JL et al (2009) Trends in blood lead levels and blood lead testing among US children aged 1 to 5 years, 1988–2004. Pediatrics 123(3):e376–e385
Kapoor SC, van Rossum GD, O’Neill KJ, Mercorella I (1985) Uptake of inorganic lead in vitro by isolated mitochondria and tissue slices of rat renal cortex. Biochem Pharmacol 34(9):1439–1448
Knowles SO, Donaldson WE (1996) Dietary lead alters fatty acid composition and membrane peroxidation in chick liver microsomes. Poult Sci 75(12):1498–1500
Krakowiak P, Goodlin-Jones B, Hertz-Picciotto I, Croen LA, Hansen RL (2008) Sleep problems in children with autism spectrum disorders, developmental delays, and typical development: a population-based study. J Sleep Res 17(2):197–206
Landrigan PJ, Todd AC (1994) Lead poisoning. West J Med 161(2):153–159
Lanphear BP (2007) The conquest of lead poisoning: a Pyrrhic victory. Environ Health Perspect 115(10):A484–A485
Lanphear BP, Hornung R, Khoury J, Yolton K, Baghurst P, Bellinger DC et al (2005) Low-level environmental lead exposure and children’s intellectual function: an international pooled analysis. Environ Health Perspect 113(7):894–899
Lawton LJ, Donaldson WE (1991) Lead-induced tissue fatty acid alterations and lipid peroxidation. Biol Trace Elem Res 28(2):83–97
Lidsky TI, Schneider JS (2003) Lead neurotoxicity in children: basic mechanisms and clinical correlates. Brain 126(Pt 1):5–19
London EA (2000) The environment as an etiologic factor in autism: a new direction for research. Environ Health Perspect 108(Suppl 3):401–404
Lord C, Rutter M, Le Couteur A (1994) Autism Diagnostic Interview-Revised: a revised version of a diagnostic interview for caregivers of individuals with possible pervasive developmental disorders. J Autism Dev Disord 24(5):659–685
Lord C, Pickles A, McLennan J, Rutter M, Bregman J, Folstein S et al (1997) Diagnosing autism: analyses of data from the Autism Diagnostic Interview. J Autism Dev Disord 27(5):501–517
Lord C, Rutter M, DiLavore PC, Risi S (2000) The autism diagnostic observation schedule (ADOS). Western Psychological Services, Los Angeles
Lord C, Rutter M, DiLavore PC, Risi S (2003) Autism diagnostic observation schedule manual. Western Psychological Services, Los Angeles
Masuda J, Takayama E, Satoh A, Ida M, Shinohara T, Kojima-Aikawa K et al (2004) Levels of annexin IV and V in the plasma of pregnant and postpartum women. Thromb Haemost 91(6):1129–1136
McBride HM, Neuspiel M, Wasiak S (2006) Mitochondria: more than just a powerhouse. Curr Biol 16(14):R551–R560
McCabe MJ Jr, Lawrence DA (1991) Lead, a major environmental pollutant, is immunomodulatory by its differential effects on CD4 + T cells subsets. Toxicol Appl Pharmacol 111(1):13–23
McCabe MJ Jr, Dias JA, Lawrence DA (1991) Lead influences translational or posttranslational regulation of Ia expression and increases invariant chain expression in mouse B cells. J Biochem Toxicol 6(4):269–276
Moser R, Oberley TD, Daggett DA, Friedman AL, Johnson JA, Siegel FL (1995) Effects of lead administration on developing rat kidney. I. Glutathione S-transferase isoenzymes. Toxicol Appl Pharmacol 131(1):85–93
Needleman H (2004a) Lead poisoning. Annu Rev Med 55:209–222
Needleman HL (2004b) Low level lead exposure and the development of children. Southeast Asian J Trop Med Public Health 35(2):252–254
Needleman H (2009) Low level lead exposure: history and discovery. Ann Epidemiol 19(4):235–238
Needleman HL, Landrigan PJ (2004) What level of lead in blood is toxic for a child? Am J Public Health 94(1):8 author reply 9
Nepom GT, Erlich H (1991) MHC class-II molecules and autoimmunity. Annu Rev Immunol 9:493–525
Oberley TD, Friedman AL, Moser R, Siegel FL (1995) Effects of lead administration on developing rat kidney. II. Functional, morphologic, and immunohistochemical studies. Toxicol Appl Pharmacol 131(1):94–107
Qu W, Diwan BA, Liu J, Goyer RA, Dawson T, Horton JL et al (2002) The metallothionein-null phenotype is associated with heightened sensitivity to lead toxicity and an inability to form inclusion bodies. Am J Pathol 160(3):1047–1056
Richardson AJ, Ross MA (2000) Fatty acid metabolism in neurodevelopmental disorder: a new perspective on associations between attention-deficit/hyperactivity disorder, dyslexia, dyspraxia and the autistic spectrum. Prostaglandins Leukot Essent Fatty Acids 63(1–2):1–9
Rossi E (2008) Low level environmental lead exposure—a continuing challenge. Clin Biochem Rev 29(2):63–70
Rossier MF (2006) T channels and steroid biosynthesis: in search of a link with mitochondria. Cell Calcium 40(2):155–164
Rubinsztein DC, Ravikumar B, Acevedo-Arozena A, Imarisio S, O’Kane CJ, Brown SD (2005) Dyneins, autophagy, aggregation and neurodegeneration. Autophagy 1(3):177–178
Singh VK, Rivas WH (2004) Prevalence of serum antibodies to caudate nucleus in autistic children. Neurosci Lett 355(1–2):53–56
Stamova B, Green PG, Tian Y, Hertz-Picciotto I, Pessah IN, Hansen R et al (submitted) Correlations between gene expression and mercury levels in blood of boys with and without autism
Struzynska L, Dabrowska-Bouta B, Koza K, Sulkowski G (2007) Inflammation-like glial response in lead-exposed immature rat brain. Toxicol Sci 95(1):156–162
Suzuki T, Morimura S, Diccianni MB, Yamada R, Hochi S, Hirabayashi M et al (1996) Activation of glutathione transferase P gene by lead requires glutathione transferase P enhancer I. J Biol Chem 271(3):1626–1632
Tang Y, Schapiro MB, Franz DN, Patterson BJ, Hickey FJ, Schorry EK et al (2004) Blood expression profiles for tuberous sclerosis complex 2, neurofibromatosis type 1, and Down’s syndrome. Ann Neurol 56(6):808–814
Virgolini MB, Chen K, Weston DD, Bauter MR, Cory-Slechta DA (2005) Interactions of chronic lead exposure and intermittent stress: consequences for brain catecholamine systems and associated behaviors and HPA axis function. Toxicol Sci 87(2):469–482
Virgolini MB, Bauter MR, Weston DD, Cory-Slechta DA (2006) Permanent alterations in stress responsivity in female offspring subjected to combined maternal lead exposure and/or stress. Neurotoxicology 27(1):11–21
Walton JR (1973) Granules containing lead in isolated mitochondria. Nature 243(5402):100–101
Walton J, Buckley IK (1977) The lead-poisoned cell: a fine structural study using cultured kidney cells. Exp Mol Pathol 27(2):167–182
Warren RP, Singh VK, Averett RE, Odell JD, Maciulis A, Burger RA et al (1996) Immunogenetic studies in autism and related disorders. Mol Chem Neuropathol 28(1–3):77–81
Warren RP, Odell JD, Warren WL, Burger RA, Maciulis A, Daniels WW et al (1997) Brief report: immunoglobulin A deficiency in a subset of autistic subjects. J Autism Dev Disord 27(2):187–192
White LD, Cory-Slechta DA, Gilbert ME, Tiffany-Castiglioni E, Zawia NH, Virgolini M et al (2007) New and evolving concepts in the neurotoxicology of lead. Toxicol Appl Pharmacol 225(1):1–27
Wiest MM, German JB, Harvey DJ, Watkins SM, Hertz-Picciotto I (2009) Plasma fatty acid profiles in autism: a case-control study. Prostaglandins Leukot Essent Fatty Acids 80(4):221–227
Wright LS, Kornguth SE, Oberley TD, Siegel FL (1998) Effects of lead on glutathione S-transferase expression in rat kidney: a dose-response study. Toxicol Sci 46(2):254–259
Yoon S, Han SS, Rana SV (2008) Molecular markers of heavy metal toxicity–a new paradigm for health risk assessment. J Environ Biol 29(1):1–14
Zhang A (2006) Advanced analysis of gene expression microarray data. World Scientific, Singapore; Hackensack, NJ
Acknowledgments
The authors thank the CHARGE study staff for recruitment, assessment, and data collection and the many families who participated. We also thank the UCD genomics core for processing the samples. This study was supported by NIH grant (I P01 ES11269) (1 R01 ES015359), the U.S. Environmental Protection Agency through the Science to ARCHIVE Results (STAR) program (R829388, R833292), a Cure Autism Now (Autism Speaks) grant, a UC Discovery (Industry-University Cooperative Research Program) grant, and an award from the Clinical and Translational Science Center (CTSC) and MIND Institute at UCD. The authors declare they have no competing financial interests.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Fig. 1
Comparison of the blood lead levels for the AU and TD subjects in this study with or without log2 transformation. Samples are shown in X-axis. The blood lead levels without log2 transformation are shown in the left Y-axis. The log2 Pb values are shown in the right Y-axis (TIFF 4805 kb)
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Tian, Y., Green, P.G., Stamova, B. et al. Correlations of Gene Expression with Blood Lead Levels in Children with Autism Compared to Typically Developing Controls. Neurotox Res 19, 1–13 (2011). https://doi.org/10.1007/s12640-009-9126-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12640-009-9126-x