Abstract
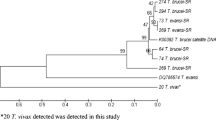
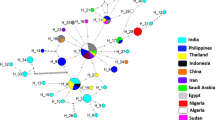
Trypanosoma vivax (sub-genus Duttonella) is largely responsible for non profitable livestock production in sub-Sahara Africa. In Nigeria, no study has addressed the molecular characteristic of T. vivax except Y486. Hence, we characterized and assessed the genetic diversity among T. vivax detected in naturally infected cattle in Nigeria using internal transcribed spacer 1 (ITS1) of ribosoma DNA (rDNA) and diagnostic antigen gene (DAG) sequences. The length of ITS1 and DAG sequences range from 215–220 to 257–338 bp, respectively and the mean G–C contents were 60 and 61.5 %. Homology search revealed 93–99 and 95–100 % homologies to T. vivax DAG and ITS1 sequences from GenBank. Aligned sequences revealed both ITS1 rDNA and DAG to be less polymorphic but DAG sequences of the Y486 strain and its clone showed marked variation from autochthonous strains. Phylogenetic analysis yielded tree that grouped T. vivax ITS1rDNA gene and DAG sequences into two main clades each. Considering the ITI1 rDNA sequences, clade A contained autochthonous T. vivax within which the South American sequences clustered, clade B contained the sequences of T. vivax from East Africa. Analysis of DAG revealed that the clade A contains autochthonous T. vivax sequences but clade B contained the Y486 and its clones. In conclusion, the diagnostic antigen gene sequences of the T. vivax detected in this study may have undergone considerable gene recombination through time and suggests that more than one strain of T. vivax exist among cattle population in Nigeria.





Similar content being viewed by others
References
Adams ER, Hamilton PB, Rodrigues AC, Malele II, Delespaux V, Teixeira MMG, Gibson W (2010) New Trypanosoma (Duttonella) vivax genotypes from tsetse flies in East Africa. Parasitology 137:641–650
Chamond N, Cosson A, Blom-Potar MC, Jouvion G, D’Archivio S, Medina M, Droin-Bergère S, Huerre M, Goyard S, Minoprio P (2010) Trypanosoma vivax infections: pushing ahead with mouse models for the study of Nagana. Parasitological, hematological and pathological parameters. PLoS Neg Trop Dis 10:e792
Cortez AP, Ventura RM, Rodrigues AC, Batista JS, Paiva F, Añez N, Machado RZ, Gibson WC, Teixeira MMG (2006) The taxonomic and phylogenetic relationships of Trypanosoma vivax from South America and Africa. Parasitology 133:159–169
Dirie EF, Croft SL, Molyneux DH (1986) Morphological changes of Trypanosomavivax in mice. Vet Parasitol 19:23–27
Dirie MF, Otte MJ, Thatthi R, Gardner PR (1993a) Comparative studies of Trypanosoma (Duttonella) vivax isolates from Colombia. Parasitology 106:21–29
Dirie MF, Murphy NB, Gardiner PR (1993b) DNA finger printing of Trypanosoma vivax isolates rapidly identifies intraspecific relationships. J Eukaryot Microbiol 40:132–134
Fasogbon AI, Knowles G, Gardiner PR (1990) A comparison of the isoenzymes of Trypanosoma (Duttonella) vivax isolates from East and West Africa. Int J Parasitol 20:389–394
Garcia H, Garcia ME, Perez H, Mendoza-Leon A (2005) The detection and PCR-based characterization of the parasites causing trypanosomiasis in water-buffalo herds in Venezuela. Ann Trop Med Parasitol 99(4):1–12
Garcia HA, Rodrigues AC, Rodrigues CMF, Bengaly Z, Minervino AHH, Riet-Correa F, Machado RZ, Paiva F, Batista JS, Neves L, Hamilton PB, Teixeira MMG (2014) Microsatellite analysis supports clonal propagation and reduced divergence of Trypanosoma vivax from asymptomatic to fatally infected livestock in South America compared to West Africa. Parasites Vectors 7:210
Gardiner PR, Mahmoud MM (1992) Salivarian trypanosomes causing disease in livestock outside sub-saharan Africa. In: Kreier JP, Baker JR (eds) Parasitic protozoa. Academic Press, London, pp 277–313
Gibson W, Peacock L, Ferris V, Fischer K, Livingstone J, Thomas J, Bailey M (2015) Genetic recombination between human and animal parasites creates novel strains of human pathogen. PLoS Negl Trop Dis 9(3):e0003665. doi:10.1371/journal.pntd.0003665
Jones TW, Davila AM (2001) Trypanosoma vivax-out of Africa. Trends Parasitol 17:99–101
Leeflang P, Buys J, Blockamp C (1976) Study on Trypanosoma vivax: infectivity and serial maintenance of natural bovine isolates in mice. Int J Parasitol 6:413–417
Malele I, Craske L, Knight C, Ferris V, Njiru Z, Hamilton P, Lehane S, Lehane M, Gibson WC (2003) The use of specific and generic primers to identify trypanosome infections of wild tsetse flies in Tanzania by PCR. Inf Gen Evol 3:271–279
Masake RA, Majiwa PAO, Moloo SK, Makau JM, Njuguna JT, Maina M, Kabata J, ole-MoiYoi OK, Nantulya VM (1997) Sensitive and specific detection of Trypanosoma vivax using the polymerase chain reaction. Exp Parasitol 85:193–205
Nakayima J, Nakao R, Alhassan A, Hayashida K, Namangala B, Mahama C, Afakye K, Sugimoto C (2013) Genetic diversity among Trypanosoma (Duttonella) vivax strains from Zambia and Ghana, based on cathepsin L-like gene. Parasite 20:24
Ng’ayo MO, Njiru ZK, Kenya EU, Muluvi GM, Osir EO, Masiga DK (2005) Detection of trypanosomes in small ruminants and pigs in western Kenya: important reservoirs in the epidemiology of sleeping sickness? Kinetoplastid Biol Dis 4:5. doi:10.1186/1475-9292-4-5
Njiru ZK, Constantine CC, Guya S, Crowther J, Kiragu JM, Thompson RCA, Davila AMR (2005) The use of ITS1 rDNA PCR in detecting pathogenic African trypanosomes. Parasitol Res 95:186–192
Osorio A, Madruga CR, Desquesnes M, Soares CO, Ribeiro LRR, da Costa SCG (2008) Trypanosoma (Duttonella) vivax: its biology, epidemiology, pathogenesis, and introduction in the new world—a review. Mem Inst Oswaldo Cruz 103:1–13
Rodrigues AC, Neves L, Garcia HA, Viola LB, Marcili A, Maia Da Silva F, Sigauque I, Batista JS, Paiva F, Teixeira MMG (2008) Phylogenetic analysis of Trypanosoma vivax supports the separation of South American/West African from East African isolates and a new T. vivax-like genotype infecting a nyala antelope from Mozambique. Parasitology 135:1317–1328
Sanni T, Onasanya G, Adefenwa MA, Yakubu A, Ikeobi CON, Adebambo OA, Talabi AO, Ozoje O, Wheto M, Takeet MI, Peters SO, De Donato M, Thomas B, Imumorin IG (2013) Molecular diagnosis of subclinical African Trypanosoma vivax infection and association with physiological indices and serum metabolites in extensively managed goats in the tropics. Open J Vet Med 3:39–45
Stevens J, Rambaut A (2001) Evolutionary rate differences in trypanosomes. Inf Gen Evol 1:143–150
Takeet MI, Fagbemi BO, De Donato M, Yakubu A, Rodulfo HE, Peters SO, Wheto M, Imumorin IG (2013) Molecular survey of pathogenic trypanosomes in naturally infected Nigerian cattle. Res Vet Sci 94(3):555–561
Tamura K, Peterson D, Peterson N, Stecher G, Nei M (2011) Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28(10):2731–2739
Acknowledgments
Financial support provided by the Educational Trust Fund of the Federal Republic of Nigeria (Grant No. TETFUND-2010) and also thankful for the financial support by the College of Agriculture and Life Sciences, Cornell University, Ithaca, NY and Zoetis, Inc. Additional support by National Research Initiative Competitive Grant Program (Grant No. 2006-35205-16864) from the USDA National Institute of Food and Agriculture; USDA-NIFA Research Agreements (Nos. 2009-65205-05635, 2010-34444-20729). We also thank the entire staff of the Department of Parasitology and Entomology, Faculty of Veterinary Medicine, Ahmadu Bello University, Zaria, Nigeria for permission granted to use their laboratory facilities for part of this study, with special gratitude to Prof. I. A. Lawal and Dr. O.O. Okubanjo.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Takeet, M.I., Fagbemi, B.O., Peters, S.O. et al. Genetic diversity among Trypanosoma vivax strains detected in naturally infected cattle in Nigeria based on ITS1 of rDNA and diagnostic antigen gene sequences. J Parasit Dis 41, 433–441 (2017). https://doi.org/10.1007/s12639-016-0822-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12639-016-0822-1