Abstract
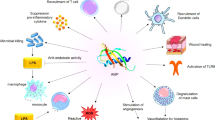
Peptides are considered very important due to the diversity expressed through their amino acid sequence, structure variation, large spectrum, and their essential role in biological systems. Antimicrobial peptides (AMPs) emerged as a potent tool in therapy owing to their antimicrobial properties but also their ability to trespass the membranes, specificity, and low toxicity. They comprise a variety of peptides from which specific amino acid-rich peptides are of interest to the current review due to their features in metal interaction and cell penetration. Histidine-rich peptides such as Histatins belong to the metal binding salivary residing peptides with efficient antibacterial, antifungal, and wound-healing activities. Furthermore, their ability to activate in acidic environment attracted the attention to their potential in therapy. The current review covers the current knowledge about AMPs and critically assess the potential of associating with metal ions both structurally and functionally. This review provides interesting hints for the advantages provided by AMPs and metal ions in biomedicine, making use of their direct properties in brain diseases therapy or in the creation of new bio-functionalized nanoparticles for cancer diagnosis and treatment.





Similar content being viewed by others
Abbreviations
- Aβ:
-
Amyloid-beta
- AD:
-
Alzheimer’s disease
- ADP:
-
Antimicrobial database
- AMPs:
-
Antimicrobial peptides
- APP:
-
Amyloid precursor protein
- BioFNPs:
-
Bio-functionalized nanoparticles
- CPPs:
-
Cell-penetrating peptides
- CPT:
-
Camptothecin
- DDSs:
-
Drug-delivery systems
- FNPs:
-
Functionalized NPs
- his1 (or htn1):
-
Histatin gene 1
- his2 (or htn2):
-
Histatin gene 2
- HSTs:
-
Histatins
- MPACs:
-
Metal-peptide attenuating compounds
- NiSOD:
-
Nickel-superoxide dismutase
- NPs:
-
Nanoparticles
- PPT:
-
Podophyllotoxin
- PTX:
-
Paclitaxel
- SAARPs:
-
Specific amino acid-rich peptides
- TBI:
-
Traumatic brain injury
- Xaa:
-
Random amino acid
References
Boman HG (2003) Antibacterial peptides: basic facts and emerging concepts. J Intern Med 254:197–215. https://doi.org/10.1046/j.1365-2796.2003.01228.x
Hancock RE, Sahl HG (2006) Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat Biotechnol 24:1551–1557. https://doi.org/10.1038/nbt1267
Zasloff M (2002) Antimicrobial peptides of multicellular organisms. Nature 415:389–395. https://doi.org/10.1038/415389a
Wang G (2010) Antimicrobial peptides: discovery, design and novel therapeutic strategies. CABI. https://doi.org/10.1079/9781845936570.0000
Menousek J, Mishra B, Hanke ML, Heim CE, Kielian T, Wang G (2012) Database screening and in vivo efficacy of antimicrobial peptides against methicillin-resistant Staphylococcus aureus USA300. Int J Antimicrob Agents 39:402–406. https://doi.org/10.1016/j.ijantimicag.2012.02.003
Wang G, Li X, Wang Z (2016) APD3: the antimicrobial peptide database as a tool for research and education. Nucleic Acids Res 44:D1087–D1093. https://doi.org/10.1093/nar/gkv1278
Wang G, Li X, Wang Z (2019) The antimicrobial peptide database (APD). Dept of Pathology & Microbiology, UNMC, Omaha http://aps.unmc.edu/AP/main.php. Accessed 03 March 2019
Gogoladze G, Grigolava M, Vishnepolsky B, Chubinidze M, Duroux P, Lefranc MP, Pirtskhalava M (2014) DBAASP: database of antimicrobial activity and structure of peptides. FEMS Microbiol Lett 357:63–68. https://doi.org/10.1111/1574-6968.12489
Zhao X, Wu H, Lu H, Li G, Huang Q (2013) LAMP: a database linking antimicrobial peptides. PLoS One 8:e66557. https://doi.org/10.1371/journal.pone.0066557
Yang X, Lee WH, Zhang Y (2012) Extremely abundant antimicrobial peptides existed in the skins of nine kinds of chinese odorous frogs. J Proteome Res 11:306–319. https://doi.org/10.1021/pr200782u
Lai Y, Gallo RL (2009) AMPed up immunity: how antimicrobial peptides have multiple roles in immune defense. Trends Immunol 30:131–141. https://doi.org/10.1016/j.it.2008.12.003
Łoboda D, Kozłowski H, Rowińska-Żyrek M (2018) Antimicrobial peptide–metal ion interactions–a potential way of activity enhancement. New J Chem 42:7560–7568. https://doi.org/10.1039/c7nj04709f
Conde R, Arguello M, Izquierdo J, Noguez R, Moreno M, Lanz H (2012) Natural antimicrobial peptides from eukaryotic organisms. In: Antimicrobial agents. Varaprasad Bobbarala, IntechOpen, Arizona, pp 51–72. https://doi.org/10.5772/33293
Bulet P, Hetru C, Dimarcq JL, Hoffmann D (1999) Antimicrobial peptides in insects; structure and function. Dev Comp Immunol 23:329–344
Dudev T, Lim C (2008) Metal binding affinity and selectivity in metalloproteins: insights from computational studies. Annu Rev Biophys 37:97–116. https://doi.org/10.1146/annurev.biophys.37.032807.125811
Harding MM, Nowicki MW, Walkinshaw MD (2010) Metals in protein structures: a review of their principal features. Crystallogr Rev 16:247–302. https://doi.org/10.1080/0889311x.2010.485616
Holm RH, Kennepohl P, Solomon EI (1996) Structural and functional aspects of metal sites in biology. Chem Rev 96:2239–2314. https://doi.org/10.1021/cr9500390
Auld DS (2001) Zinc coordination sphere in biochemical zinc sites. Springer, Netherlands. https://doi.org/10.1007/978-94-017-3728-9_6
Kulon K, Valensin D, Kamysz W, Valensin G, Nadolski P, Porciatti E, Gaggelli E, Kozłowski H (2008) The His-His sequence of the antimicrobial peptide demegen P-113 makes it very attractive ligand for Cu2+. J Inorg Biochem 102:960–972. https://doi.org/10.1016/j.jinorgbio.2007.12.021
Mishra B, Wang G (2012) The importance of amino acid composition in natural AMPs: an evolutional, structural, and functional perspective. Front Immunol 3:221. https://doi.org/10.3389/fimmu.2012.00221
Wang G, Li X, Wang Z (2009) APD2: the updated antimicrobial peptide database and its application in peptide design. Nucleic Acids Res 37:D933–D937. https://doi.org/10.1093/nar/gkn823
Wang Z, Wang G (2004) APD: the antimicrobial peptide database. Nucleic Acids Res 32:D590–D592. https://doi.org/10.1093/nar/gkh025
Wang G, Watson KM, Peterkofsky A, Buckheit RW Jr (2010) Identification of novel human immunodeficiency virus type 1-inhibitory peptides based on the antimicrobial peptide database. Antimicrob Agents Chemother 54:1343–1346. https://doi.org/10.1128/AAC.01448-09
Miteva M, Andersson M, Karshikoff A, Otting G (1999) Molecular electroporation: a unifying concept for the description of membrane pore formation by antibacterial peptides, exemplified with NK-lysin. FEBS Lett 462:155–158. https://doi.org/10.1016/s0014-5793(99)01520-3
Pokorny A, Almeida PF (2004) Kinetics of dye efflux and lipid flip-flop induced by delta-lysin in phosphatidylcholine vesicles and the mechanism of graded release by amphipathic, alpha-helical peptides. Biochemistry 43:8846–8857. https://doi.org/10.1021/bi0497087
Pokorny A, Almeida PF (2005) Permeabilization of raft-containing lipid vesicles by delta-lysin: a mechanism for cell sensitivity to cytotoxic peptides. Biochemistry 44:9538–9544. https://doi.org/10.1021/bi0506371
Chan DI, Prenner EJ, Vogel HJ (2006) Tryptophan- and arginine-rich antimicrobial peptides: structures and mechanisms of action. Biochim Biophys Acta 1758:1184–1202. https://doi.org/10.1016/j.bbamem.2006.04.006
Tieleman DP (2004) The molecular basis of electroporation. BMC Biochem 5:10. https://doi.org/10.1186/1471-2091-5-10
Brogden KA (2005) Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat Rev Microbiol 3:238–250. https://doi.org/10.1038/nrmicro1098
Xing G, DeRose VJ (2001) Designing metal–peptide models for protein structure and function. Curr Opin Chem Biol 5:196–200. https://doi.org/10.1016/s1367-5931(00)00190-3
Licini G, Scrimin P (2003) Metal-ion-binding peptides: from catalysis to protein tagging. Angew Chem Int Ed Engl 42:4572–4575. https://doi.org/10.1002/anie.200301668
Turk BE, Cantley LC (2003) Peptide libraries: at the crossroads of proteomics and bioinformatics. Curr Opin Chem Biol 7:84–90. https://doi.org/10.1016/s1367-5931(02)00004-2
Berkessel A, Hérault DA (1999) Discovery of peptide-zirconium complexes that mediate phosphate hydrolysis by batch screening of a combinatorial undecapeptide library. Angew Chem Int Ed 38:102–105. https://doi.org/10.1002/(sici)1521-3773(19990115)38:1/2<102::aid-anie102>3.0.co;2-h
Huang X, Pieczko ME, Long EC (1999) Combinatorial optimization of the DNA cleaving Ni(II) x Xaa-Xaa-His metallotripeptide domain. Biochemistry 38:2160–2166. https://doi.org/10.1021/bi982587o
Barondeau DP, Kassmann CJ, Bruns CK, Tainer JA, Getzoff ED (2004) Nickel superoxide dismutase structure and mechanism. Biochemistry 43:8038–8047. https://doi.org/10.1021/bi0496081
Wuerges J, Lee JW, Yim YI, Yim HS, Kang SO, Djinovic Carugo K (2004) Crystal structure of nickel-containing superoxide dismutase reveals another type of active site. Proc Natl Acad Sci U S A 101:8569–8574. https://doi.org/10.1073/pnas.0308514101
Garcia J, Gerber SH, Sugita S, Sudhof TC, Rizo J (2004) A conformational switch in the piccolo C2A domain regulated by alternative splicing. Nat Struct Mol Biol 11:45–53. https://doi.org/10.1038/nsmb707
Barondeau DP, Getzoff ED (2004) Structural insights into protein-metal ion partnerships. Curr Opin Struct Biol 14:765–774. https://doi.org/10.1016/j.sbi.2004.10.012
Chimento DP, Mohanty AK, Kadner RJ, Wiener MC (2003) Substrate-induced transmembrane signaling in the cobalamin transporter BtuB. Nat Struct Biol 10:394–401. https://doi.org/10.1038/nsb914
Adams TE, Hockin MF, Mann KG, Everse SJ (2004) The crystal structure of activated protein C-inactivated bovine factor Va: implications for cofactor function. Proc Natl Acad Sci U S A 101:8918–8923. https://doi.org/10.1073/pnas.0403072101
Wilson JJ, Matsushita O, Okabe A, Sakon J (2003) A bacterial collagen-binding domain with novel calcium-binding motif controls domain orientation. EMBO J 22:1743–1752. https://doi.org/10.1093/emboj/cdg172
Conklin SE, Bridgman EC, Su Q, Riggs-Gelasco P, Haas KL, Franz KJ (2017) Specific histidine residues confer histatin peptides with copper-dependent activity against Candida albicans. Biochemistry 56:4244–4255. https://doi.org/10.1021/acs.biochem.7b00348
de Sousa-Pereira P, Amado F, Abrantes J, Ferreira R, Esteves PJ, Vitorino R (2013) An evolutionary perspective of mammal salivary peptide families: cystatins, histatins, statherin and PRPs. Arch Oral Biol 58:451–458. https://doi.org/10.1016/j.archoralbio.2012.12.011
Imamura Y, Fujigaki Y, Oomori Y, Ouryouji K, Yanagisawa S, Miyazawa H, Wang PL (2009) Transcriptional regulation of the salivary histatin gene: finding of a strong positive regulatory element and its binding protein. J Biochem 145:279–288. https://doi.org/10.1093/jb/mvn165
Troxler RF, Offner GD, Xu T, Vanderspek JC, Oppenheim FG (1990) Structural relationship between human salivary histatins. J Dent Res 69:2–6. https://doi.org/10.1177/00220345900690010101
Sun X, Salih E, Oppenheim FG, Helmerhorst EJ (2009) Kinetics of histatin proteolysis in whole saliva and the effect on bioactive domains with metal-binding, antifungal, and wound-healing properties. FASEB J 23:2691–2701. https://doi.org/10.1096/fj.09-131045
Khurshid Z, Najeeb S, Mali M, Moin SF, Raza SQ, Zohaib S, Sefat F, Zafar MS (2017) Histatin peptides: pharmacological functions and their applications in dentistry. Saudi Pharm J 25:25–31. https://doi.org/10.1016/j.jsps.2016.04.027
Melino S, Rufini S, Sette M, Morero R, Grottesi A, Paci M, Petruzzelli R (1999) Zn(2+) ions selectively induce antimicrobial salivary peptide histatin-5 to fuse negatively charged vesicles. Identification and characterization of a zinc-binding motif present in the functional domain. Biochemistry 38:9626–9633. https://doi.org/10.1021/bi990212c
Grogan J, McKnight CJ, Troxler RF, Oppenheim FG (2001) Zinc and copper bind to unique sites of histatin 5. FEBS Lett 491:76–80. https://doi.org/10.1016/s0014-5793(01)02157-3
Puri S, Edgerton M (2014) How does it kill?: understanding the candidacidal mechanism of salivary histatin 5. Eukaryot Cell 13:958–964. https://doi.org/10.1128/EC.00095-14
Wiesner J, Vilcinskas A (2010) Antimicrobial peptides: the ancient arm of the human immune system. Virulence 1:440–464. https://doi.org/10.4161/viru.1.5.12983
Helmerhorst EJ, Breeuwer P, van ‘t Hof W, Walgreen-Weterings E, Oomen LCJM, Veerman ECI, Amerongen AVN, Abee T (1999) The cellular target of histatin 5 on Candida albicans is the energized mitochondrion. J Biol Chem 274:7286–7291. https://doi.org/10.1074/jbc.274.11.7286
Li XS, Reddy MS, Baev D, Edgerton M (2003) Candida albicans Ssa1/2p is the cell envelope binding protein for human salivary histatin 5. J Biol Chem 278:28553–28561. https://doi.org/10.1074/jbc.M300680200
Helmerhorst EJ, Troxler RF, Oppenheim FG (2001) The human salivary peptide histatin 5 exerts its antifungal activity through the formation of reactive oxygen species. Proc Natl Acad Sci U S A 98:14637–14642. https://doi.org/10.1073/pnas.141366998
Sankararamakrishnan R, Verma S, Kumar S (2005) ATCUN-like metal-binding motifs in proteins: identification and characterization by crystal structure and sequence analysis. Proteins 58:211–221. https://doi.org/10.1002/prot.20265
Harford C, Sarkar B (1997) Amino terminal Cu(II)- and Ni(II)-binding (ATCUN) motif of proteins and peptides: metal binding, DNA cleavage, and other properties. Acc Chem Res 30:123–130. https://doi.org/10.1021/ar9501535
Alies B, Badei B, Faller P, Hureau C (2012) Reevaluation of copper(I) affinity for amyloid-beta peptides by competition with ferrozine–an unusual copper(I) indicator. Chemistry (Easton) 18:1161–1167. https://doi.org/10.1002/chem.201102746
Gusman H, Lendenmann U, Grogan J, Troxler RF, Oppenheim FG (2001) Is salivary histatin 5 a metallopeptide? Biochim Biophys Acta Protein Struct Mol Enzymol 1545:86–95. https://doi.org/10.1016/s0167-4838(00)00265-x
Puri S, Li R, Ruszaj D, Tati S, Edgerton M (2015) Iron binding modulates candidacidal properties of salivary histatin 5. J Dent Res 94:201–208. https://doi.org/10.1177/0022034514556709
Kurowska E, Bonna A, Goch G, Bal W (2011) Salivary histatin-5, a physiologically relevant ligand for Ni(II) ions. J Inorg Biochem 105:1220–1225. https://doi.org/10.1016/j.jinorgbio.2011.06.002
Melino S, Santone C, Di Nardo P, Sarkar B (2014) Histatins: salivary peptides with copper(II)- and zinc(II)-binding motifs: perspectives for biomedical applications. FEBS J 281:657–672. https://doi.org/10.1111/febs.12612
Melino S, Gallo M, Trotta E, Mondello F, Paci M, Petruzzelli R (2006) Metal-binding and nuclease activity of an antimicrobial peptide analogue of the salivary histatin 5. Biochemistry 45:15373–15383. https://doi.org/10.1021/bi0615137
Tay WM, Hanafy AI, Angerhofer A, Ming LJ (2009) A plausible role of salivary copper in antimicrobial activity of histatin-5–metal binding and oxidative activity of its copper complex. Bioorg Med Chem Lett 19:6709–6712. https://doi.org/10.1016/j.bmcl.2009.09.119
Porciatti E, Milenkovic M, Gaggelli E, Valensin G, Kozlowski H, Kamysz W, Valensin D (2010) Structural characterization and antimicrobial activity of the Zn(II) complex with P113 (demegen), a derivative of histatin 5. Inorg Chem 49:8690–8698. https://doi.org/10.1021/ic902547t
Cobine PA, Ojeda LD, Rigby KM, Winge DR (2004) Yeast contain a non-proteinaceous pool of copper in the mitochondrial matrix. J Biol Chem 279:14447–14455. https://doi.org/10.1074/jbc.M312693200
Chari RV (2008) Targeted cancer therapy: conferring specificity to cytotoxic drugs. Acc Chem Res 41:98–107. https://doi.org/10.1021/ar700108g
Monje M, Dietrich J (2012) Cognitive side effects of cancer therapy demonstrate a functional role for adult neurogenesis. Behav Brain Res 227:376–379. https://doi.org/10.1016/j.bbr.2011.05.012
Monsuez JJ, Charniot JC, Vignat N, Artigou JY (2010) Cardiac side-effects of cancer chemotherapy. Int J Cardiol 144:3–15. https://doi.org/10.1016/j.ijcard.2010.03.003
Dinca A, Chien WM, Chin MT (2016) Intracellular delivery of proteins with cell-penetrating peptides for therapeutic uses in human disease. Int J Mol Sci 17:263. https://doi.org/10.3390/ijms17020263
Helmfors H, Eriksson J, Langel U (2015) Optimized luciferase assay for cell-penetrating peptide-mediated delivery of short oligonucleotides. Anal Biochem 484:136–142. https://doi.org/10.1016/j.ab.2015.05.023
Huang Y, Jiang Y, Wang H, Wang J, Shin MC, Byun Y, He H, Liang Y, Yang VC (2013) Curb challenges of the “Trojan horse” approach: smart strategies in achieving effective yet safe cell-penetrating peptide-based drug delivery. Adv Drug Deliv Rev 65:1299–1315. https://doi.org/10.1016/j.addr.2012.11.007
Kato T, Yamashita H, Misawa T, Nishida K, Kurihara M, Tanaka M, Demizu Y, Oba M (2016) Plasmid DNA delivery by arginine-rich cell-penetrating peptides containing unnatural amino acids. Bioorg Med Chem 24:2681–2687. https://doi.org/10.1016/j.bmc.2016.04.031
Fonseca SB, Pereira MP, Kelley SO (2009) Recent advances in the use of cell-penetrating peptides for medical and biological applications. Adv Drug Deliv Rev 61:953–964. https://doi.org/10.1016/j.addr.2009.06.001
Vives E, Schmidt J, Pelegrin A (2008) Cell-penetrating and cell-targeting peptides in drug delivery. Biochim Biophys Acta 1786:126–138. https://doi.org/10.1016/j.bbcan.2008.03.001
Davoudi Z, Akbarzadeh A, Rahmatiyamchi M, Movassaghpour AA, Alipour M, Nejati-Koshki K, Sadeghi Z, Dariushnejad H, Zarghami N (2014) Molecular target therapy of AKT and NF-kB signaling pathways and multidrug resistance by specific cell penetrating inhibitor peptides in HL-60 cells. Asian Pac J Cancer Prev 15:4353–4358. https://doi.org/10.7314/apjcp.2014.15.10.4353
Vargas JR, Stanzl EG, Teng NN, Wender PA (2014) Cell-penetrating, guanidinium-rich molecular transporters for overcoming efflux-mediated multidrug resistance. Mol Pharm 11:2553–2565. https://doi.org/10.1021/mp500161z
Dubikovskaya EA, Thorne SH, Pillow TH, Contag CH, Wender PA (2008) Overcoming multidrug resistance of small-molecule therapeutics through conjugation with releasable octaarginine transporters. Proc Natl Acad Sci U S A 105:12128–12133. https://doi.org/10.1073/pnas.0805374105
Lindgren M, Rosenthal-Aizman K, Saar K, Eiríksdóttir E, Jiang Y, Sassian M, Östlund P, Hällbrink M, Langel Ü (2006) Overcoming methotrexate resistance in breast cancer tumour cells by the use of a new cell-penetrating peptide. Biochem Pharmacol 71:416–425. https://doi.org/10.1016/j.bcp.2005.10.048
Fei L, Yap LP, Conti PS, Shen WC, Zaro JL (2014) Tumor targeting of a cell penetrating peptide by fusing with a pH-sensitive histidine-glutamate co-oligopeptide. Biomaterials 35:4082–4087. https://doi.org/10.1016/j.biomaterials.2014.01.047
Jiang T, Zhang Z, Zhang Y, Lv H, Zhou J, Li C, Hou L, Zhang Q (2012) Dual-functional liposomes based on pH-responsive cell-penetrating peptide and hyaluronic acid for tumor-targeted anticancer drug delivery. Biomaterials 33:9246–9258. https://doi.org/10.1016/j.biomaterials.2012.09.027
Lee ES, Gao Z, Bae YH (2008) Recent progress in tumor pH targeting nanotechnology. J Control Release 132:164–170. https://doi.org/10.1016/j.jconrel.2008.05.003
Shi K, Li J, Cao Z, Yang P, Qiu Y, Yang B, Wang Y, Long Y, Liu Y, Zhang Q, Qian J, Zhang Z, Gao H, He Q (2015) A pH-responsive cell-penetrating peptide-modified liposomes with active recognizing of integrin alphavbeta3 for the treatment of melanoma. J Control Release 217:138–150. https://doi.org/10.1016/j.jconrel.2015.09.009
Cardone RA, Casavola V, Reshkin SJ (2005) The role of disturbed pH dynamics and the Na+/H+ exchanger in metastasis. Nat Rev Cancer 5:786–795. https://doi.org/10.1038/nrc1713
Jahde E, Rajewsky MF, Baumgartl H (1982) pH distributions in transplanted neural tumors and normal tissues of BDIX rats as measured with pH microelectrodes. Cancer Res 42:1498–1504
Tannock IF, Rotin D (1989) Acid pH in tumors and its potential for therapeutic exploitation. Cancer Res 49:4373–4384
Liang J, Wu WL, Xu XD, Zhuo RX, Zhang XZ (2014) pH responsive micelle self-assembled from a new amphiphilic peptide as anti-tumor drug carrier. Colloids Surf B: Biointerfaces 114:398–403. https://doi.org/10.1016/j.colsurfb.2013.10.037
Futaki S, Nakase I, Tadokoro A, Takeuchi T, Jones AT (2007) Arginine-rich peptides and their internalization mechanisms. Biochem Soc Trans 35:784–787. https://doi.org/10.1042/BST0350784
Guo Z, Peng H, Kang J, Sun D (2016) Cell-penetrating peptides: possible transduction mechanisms and therapeutic applications. Biomedical reports 4:528–534. https://doi.org/10.3892/br.2016.639
Ziegler A (2008) Thermodynamic studies and binding mechanisms of cell-penetrating peptides with lipids and glycosaminoglycans. Adv Drug Deliv Rev 60:580–597. https://doi.org/10.1016/j.addr.2007.10.005
Zaro JL, Fei L, Shen WC (2012) Recombinant peptide constructs for targeted cell penetrating peptide-mediated delivery. J Control Release 158:357–361. https://doi.org/10.1016/j.jconrel.2012.01.039
Zhang W, Song J, Zhang B, Liu L, Wang K, Wang R (2011) Design of acid-activated cell penetrating peptide for delivery of active molecules into cancer cells. Bioconjug Chem 22:1410–1415. https://doi.org/10.1021/bc200138d
Yao J, Ma Y, Zhang W, Li L, Zhang Y, Zhang L, Liu H, Ni J, Wang R (2017) Design of new acid-activated cell-penetrating peptides for tumor drug delivery. PeerJ 5:e3429. https://doi.org/10.7717/peerj.3429
De M, Ghosh PS, Rotello VM (2008) Applications of nanoparticles in biology. Adv Mater 20:4225–4241. https://doi.org/10.1002/adma.200703183
Jiang Z, Le NDB, Gupta A, Rotello VM (2015) Cell surface-based sensing with metallic nanoparticles. Chem Soc Rev 44:4264–4274. https://doi.org/10.1039/c4cs00387j
Morales-Avila E, Ferro-Flores G, Ocampo-García BE, López-Téllez G, López-Ortega J, Rogel-Ayala DG, Sánchez-Padilla D (2017) Antibacterial efficacy of gold and silver nanoparticles functionalized with the ubiquicidin (29–41) antimicrobial peptide. J Nanomater 2017:1–10. https://doi.org/10.1155/2017/5831959
Rajchakit U, Sarojini V (2017) Recent developments in antimicrobial-peptide-conjugated gold nanoparticles. Bioconjug Chem 28:2673–2686. https://doi.org/10.1021/acs.bioconjchem.7b00368
Masood F (2016) Polymeric nanoparticles for targeted drug delivery system for cancer therapy. Mater Sci Eng C Mater Biol Appl 60:569–578. https://doi.org/10.1016/j.msec.2015.11.067
Sousa MH, Rubim JC, Sobrinho PG, Tourinho FA (2001) Biocompatible magnetic fluid precursors based on aspartic and glutamic acid modified maghemite nanostructures. J Magn Magn Mater 225:67–72. https://doi.org/10.1016/S0304-8853(00)01229-4
Tie SL, Lin YQ, Lee HC, Bae YS, Lee CH (2006) Amino acid-coated nano-sized magnetite particles prepared by two-step transformation. Colloids Surf A Physicochem Eng Asp 273:75–83. https://doi.org/10.1016/j.colsurfa.2005.08.027
Viota JL, Arroyo FJ, Delgado AV, Horno J (2010) Electrokinetic characterization of magnetite nanoparticles functionalized with amino acids. J Colloid Interface Sci 344:144–149. https://doi.org/10.1016/j.jcis.2009.11.061
Wampler HP, Ivanisevic A (2009) Nanoindentation of gold nanoparticles functionalized with proteins. Micron 40:444–448. https://doi.org/10.1016/j.micron.2009.01.002
Koh I, Wang X, Varughese B, Isaacs L, Ehrman SH, English DS (2006) Magnetic iron oxide nanoparticles for biorecognition: evaluation of surface coverage and activity. J Phys Chem B 110:1553–1558. https://doi.org/10.1021/jp0556310
Chen W, Shen HB, Li XY, Jia NQ, Xu JM (2006) Synthesis of immunomagnetic nanoparticles and their application in the separation and purification of CD34(+) hematopoietic stem cells. Appl Surf Sci 253:1762–1769. https://doi.org/10.1016/j.apsusc.2006.03.012
von Zur MC et al (2007) Superparamagnetic iron oxide binding and uptake as imaged by magnetic resonance is mediated by the integrin receptor mac-1 (CD11b/CD18): implications on imaging of atherosclerotic plaques. Atherosclerosis 193:102–111. https://doi.org/10.1016/j.atherosclerosis.2006.08.048
Li L, Wartchow CA, Danthi SN, Shen Z, Dechene N, Pease J, Choi HS, Doede T, Chu P, Ning S, Lee DY, Bednarski MD, Knox SJ (2004) A novel antiangiogenesis therapy using an integrin antagonist or anti-Flk-1 antibody coated 90Y-labeled nanoparticles. Int J Radiat Oncol Biol Phys 58:1215–1227. https://doi.org/10.1016/j.ijrobp.2003.10.057
Dharap SS, Qiu B, Williams GC, Sinko P, Stein S, Minko T (2003) Molecular targeting of drug delivery systems to ovarian cancer by BH3 and LHRH peptides. J Control Release 91:61–73. https://doi.org/10.1016/s0168-3659(03)00209-8
Missailidis S, Thomaidou D, Borbas KE, Price MR (2005) Selection of aptamers with high affinity and high specificity against C595, an anti-MUC1 IgG3 monoclonal antibody, for antibody targeting. J Immunol Methods 296:45–62. https://doi.org/10.1016/j.jim.2004.10.011
Mitra A, Mulholland J, Nan A, McNeill E, Ghandehari H, Line BR (2005) Targeting tumor angiogenic vasculature using polymer-RGD conjugates. J Control Release 102:191–201. https://doi.org/10.1016/j.jconrel.2004.09.023
Cui H, Webber MJ, Stupp SI (2010) Self-assembly of peptide amphiphiles: from molecules to nanostructures to biomaterials. Biopolymers 94:1–18. https://doi.org/10.1002/bip.21328
Song SJ, Lee S, Ryu KS, Choi JS (2017) Amphiphilic peptide nanorods based on oligo-phenylalanine as a biocompatible drug carrier. Bioconjug Chem 28:2266–2276. https://doi.org/10.1021/acs.bioconjchem.7b00247
de la Rica R, Matsui H (2010) Applications of peptide and protein-based materials in bionanotechnology. Chem Soc Rev 39:3499–3509. https://doi.org/10.1039/b917574c
Wang F, Wang Y, Zhang X, Zhang W, Guo S, Jin F (2014) Recent progress of cell-penetrating peptides as new carriers for intracellular cargo delivery. J Control Release 174:126–136. https://doi.org/10.1016/j.jconrel.2013.11.020
Chiu LS, Anderton RS, Knuckey NW, Meloni BP (2017) Peptide pharmacological approaches to treating traumatic brain injury: a case for arginine-rich peptides. Mol Neurobiol 54:7838–7857. https://doi.org/10.1007/s12035-016-0287-3
Sundman MH, Hall EE, Chen NK (2014) Examining the relationship between head trauma and neurodegenerative disease: a review of epidemiology, pathology and neuroimaging techniques. J Alzheimers Dis Parkinsonism 4:137. https://doi.org/10.4172/2161-0460.1000137
Lakatos A, Gyurcsik B, Nagy NV, Csendes Z, Weber E, Fulop L, Kiss T (2012) Histidine-rich branched peptides as Cu(II) and Zn(II) chelators with potential therapeutic application in Alzheimer’s disease. Dalton Trans 41:1713–1726. https://doi.org/10.1039/c1dt10989h
Lin CJ, Huang HC, Jiang ZF (2010) Cu(II) interaction with amyloid-beta peptide: a review of neuroactive mechanisms in AD brains. Brain Res Bull 82:235–242. https://doi.org/10.1016/j.brainresbull.2010.06.003
Sarell CJ, Wilkinson SR, Viles JH (2010) Substoichiometric levels of Cu2+ ions accelerate the kinetics of fiber formation and promote cell toxicity of amyloid-{beta} from Alzheimer disease. J Biol Chem 285:41533–41540. https://doi.org/10.1074/jbc.M110.171355
Nhan HS, Chiang K, Koo EH (2015) The multifaceted nature of amyloid precursor protein and its proteolytic fragments: friends and foes. Acta Neuropathol 129:1–19. https://doi.org/10.1007/s00401-014-1347-2
Goodman Y, Mattson MP (1994) Secreted forms of beta-amyloid precursor protein protect hippocampal neurons against amyloid beta-peptide-induced oxidative injury. Exp Neurol 128:1–12. https://doi.org/10.1006/exnr.1994.1107
Corrigan F, Thornton E, Roisman LC, Leonard AV, Vink R, Blumbergs PC, van den Heuvel C, Cappai R (2014) The neuroprotective activity of the amyloid precursor protein against traumatic brain injury is mediated via the heparin binding site in residues 96-110. J Neurochem 128:196–204. https://doi.org/10.1111/jnc.12391
Plummer SL, Corrigan F, Thornton E, Woenig JA, Vink R, Cappai R, Van Den Heuvel C (2018) The amyloid precursor protein derivative, APP96-110, is efficacious following intravenous administration after traumatic brain injury. PLoS One 13:e0190449. https://doi.org/10.1371/journal.pone.0190449
Plummer S, Van den Heuvel C, Thornton E, Corrigan F, Cappai R (2016) The neuroprotective properties of the amyloid precursor protein following traumatic brain injury. Aging Dis 7:163–179. https://doi.org/10.14336/AD.2015.0907
Huang X, Atwood CS, Moir RD, Hartshorn MA, Vonsattel JP, Tanzi RE, Bush AI (1997) Zinc-induced Alzheimer’s Abeta1-40 aggregation is mediated by conformational factors. J Biol Chem 272:26464–26470
Ginotra YP, Ramteke SN, Srikanth R, Kulkarni PP (2012) Mass spectral studies reveal the structure of Abeta1-16-Cu2+ complex resembling ATCUN motif. Inorg Chem 51:7960–7962. https://doi.org/10.1021/ic301244x
Soscia SJ, Kirby JE, Washicosky KJ, Tucker SM, Ingelsson M, Hyman B, Burton MA, Goldstein LE, Duong S, Tanzi RE, Moir RD (2010) The Alzheimer’s disease-associated amyloid beta-protein is an antimicrobial peptide. PLoS One 5:e9505. https://doi.org/10.1371/journal.pone.0009505
Huang Y, Huang J, Chen Y (2010) Alpha-helical cationic antimicrobial peptides: relationships of structure and function. Protein Cell 1:143–152. https://doi.org/10.1007/s13238-010-0004-3
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of Interests
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Moulahoum, H., Ghorbani Zamani, F., Timur, S. et al. Metal Binding Antimicrobial Peptides in Nanoparticle Bio-functionalization: New Heights in Drug Delivery and Therapy. Probiotics & Antimicro. Prot. 12, 48–63 (2020). https://doi.org/10.1007/s12602-019-09546-5
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12602-019-09546-5