Abstract
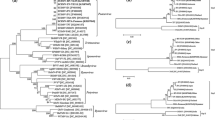
Sugarcane mosaic virus (SCMV) one of the causative viruses of mosaic disease in sugarcane occurs in sugarcane growing countries worldwide. India is the second largest sugarcane producing country and genome of SCMV from India has not been characterized so far. Hence detailed studies were carried out to characterize the virus isolates based on its complete genome. Comparative genome analyses of five new isolates were performed with previously reported SCMV full genome sequences of isolates infecting sugarcane, maize, sorghum and Canna. Sequence identity matrix and phylogenetic analyses clearly represented that Indian isolates are closely related to sugarcane infecting isolates reported from Australia, Argentina, China and Iran and they diverged as a separate subgroup from other reported maize infecting isolates from Mexico, China, Ohio, Spain, Germany, Iran, Ethiopia, Kenya and Eucador. Selection pressure analysis clearly depicted the predomination of strong purifying selection throughout the viral genome, and strongest in CI and HC-Pro gene. Evidence for few positively selected sites was identified in all the cistrons except in 6 K1 and Nib rep. Among the genomic region, CI gene has exhibited comparatively more recombination hotspots followed by HC-Pro unlike other reported isolates. As the cultivation of sugarcane first originated in India, our results from the recombination events strongly suggest that Indian SCMV populations contribute for the emergence of upcoming new recombinant SCMV isolates not only within the sugarcane isolates but also with maize infecting isolates of SCMV in other countries irrespective of geographic origin and host type.






Similar content being viewed by others
References
Achon, M. A., Serrano, L., Alonso-Duenas, N., & Porta, C. (2007). Complete genome sequences of Maize dwarf mosaic and Sugarcane mosaic virus isolates coinfecting maize in Spain. Archives of Virology, 152, 2073–2078.
Adams, I. P., Harju, V. A., Hodges, T., Hany, U., Skelton, A., Rai, S., Deka, M. K., Smith, J., Fox, A., Uzayisenga, B., Ngaboyisonga, C., Uwumukiza, B., Rutikanga, A., Rutherford, M., Ricthis, B., Phiri, N., & Boonham, N. (2014). First report of maize lethal necrosis disease in Rwanda. New Disease Reports, 29, 22.
Barber, C. A. (1921). The mosaic mottling disease of the sugarcane. International Sugar Journal, 23, 12–19.
Bernreiter, A., Garcia Teijeiro, R., Jarrin, D., Garrido, P., & Ramos, L. (2017). First report of Maize yellow mosaic virus infecting maize in Ecuador. New Disease Reports, 36, 11.
Blanc, S., Ammar, E. D., Garcıa-Lampasona, S., Dolja, V. V., Llave, C., Baker, J., & Pirone, T. P. (1998). Mutations in the potyvirus helper component protein: Effects on interactions with virions and aphid stylets. Journal of General Virology, 79, 3119–3122.
Cavatorta, J. R., Savage, A. E., Yeam, I., Gray, S. M., & Jahn, M. M. (2008). Positive Darwinian selection at single amino acid sites conferring plant virus resistance. Journal of Molecular Evolution, 67, 551–559.
Chaves-Bedoya, G., Espejel, F., Alcala-Briseno, R. I., Hernandez-Vela, J., & Silva-Rosales, L. (2011). Short distance movement of genomic negative strands in a host and nonhost for sugarcane mosaic virus (SCMV). Virology Journal, 8, 15.
Chen, J., Chen, J., & Adams, M. (2002). Characterisation of potyviruses from sugarcane and maize in China. Archives of Virology, 147, 1237–1246.
Chona, B. L., & Rafay, S. A. (1950). Studies on the sugarcane disease in India. Indian Journal of Agricultural Sciences, 20, 39–50.
Costa, A.S., &Muller, G.W.(1982). General evaluation of the impacts of viral diseases of economic crops on the development of Latin American countries. Proceedings of the first international conference on the impact of viral diseases in developing Latin American and Carribbean countries, Rio De Janeiro.
Daniels, J., & Roach, B. T. (1987). Taxonomy and evolution. In J. Heinz (Ed.), Sugarcane improvement through breeding (Vol. 1, pp. 7–84). Amsterdam: Elsevier.
Delport, W., Poon, A., Frost, S. D., & Kosakovsky Pond, S. L. (2010). Datamonkey: A suite of phylogenetic analysis tools for evolutionary biology. Bioinformatics, 26, 2455–2457.
Etherington, G. J., Dicks, J., & Roberts, I. N. (2005). Recombination analysis tool (RAT): A program for the high-throughput detection of recombination. Bioinformatics, 21, 278–281.
Fan, Z. F., Chen, H. Y., Liang, X. M., & Li, H. F. (2003). Complete sequence of the genomic RNA of the prevalent strain of a potyvirus infecting maize in China. Archives of Virology, 148, 773–782.
Fauconnier, R. (1993). Sugar cane (pp. 1–40). UK: Macmillan Press Ltd London.
Gao, B., Cui, X. W., Li, X. D., Zhang, C. Q., & Miao, H. Q. (2011). Complete genomic sequence analysis of a highly virulent isolate revealed a novel strain of sugarcane mosaic virus. Virus Genes, 43, 390–397.
Goodman, B.S. (1999). A study of South African strains of sugarcane mosaic potyvirus (SCMV) identified by sequence analysis of the 5’ region of the coat protein gene. M.Sc thesis, Department of Biotechnology, Durban University of Technology, Durban, South Africa, 136 pp.
Grisham, M.P. (2000). A guide to sugarcane diseases. CIRAD-ISSCT, CIRAD publication services, Montpellier. (pp. 249–254).
Hall, T. A. (1999). BioEdit: A user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symposium Series, 41, 95–98.
Hughes, A. L., & Hughes, M. A. (2005). Patterns of nucleotide difference in overlapping and non-overlapping reading frames of papillomavirus genomes. Virus Research, 113, 81–88.
Hughes, A. L., & Hughes, M. A. (2007). Coding sequence polymorphism in avian mitochondrial genomes reflects population histories. Molecular Ecology, 16, 1369–1376.
Koike, H., Gillespie, A. G.(1989). Mosaic. In Ricaud, BT Egan, AG Gillespie, CG Hughes (Ed.), Disease of sugarcane – major diseases (pp. 301–322).
Kong, P., & Steinbiss, H. H. (1998). Complete nucleotide sequence and analysis of the putative polyprotein of maize dwarf mosaic virus genomic RNA (Bulgarian isolate). Archives of Virology, 143, 1791–1799.
Kumar, S., Stecher, G., Li, M., Knyas, C., & Tamura, K. (2018). Mega X: Molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution, 35, 1574–1549.
Lai, M. M. C. (1992). RNA recombination in animal and plant viruses. Microbiological Reviews, 56, 61–79.
Li, L., Wang, X., & Zhou, G. (2007). Analyses of maize embryo invasion by Sugarcane mosaic virus. Plant Science, 172, 131–138.
Mahuku, G., Wangai, A., Sadessa, K., Teklewold, A., Wegary, D., Ayalneh, D., Adams, I., Smith, J., Bottomley, E., Bryce, S., Braidwood, L., Feyissa, B., Regassa, B., Wanjala, B., Kimunye, J. N., Mugambi, C., Monjero, K., & Prasanna, M. (2015). First Report of maize chlorotic mottle virus and maize lethal necrosis on Maize in Ethiopia. Plant Disease, 99, 1870.
Mangrauthia, S. K., Parameswari, B., Jain, R. K., & Praveen, S. (2008). Role of genetic recombination in the molecular architecture of Papaya ringspot virus. Biochemical Genetics, 46, 835–846.
Martin, D. P., Murrell, B., Michael, G., Khoosal, A., & Muhire, B. (2015). RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evolution, 1(1), vev003. https://doi.org/10.1093/ve/vev003.
Moradi, Z., Mehrvar, M., Nazifi, E., & Mohammad, Z. (2016). The complete genome sequences of two naturally occurring recombinant isolates of Sugarcane mosaic virus from Iran. Virus Genes, 52, 270–280.
Moury, B., Morel, C., Johansen, E., & Jacquemond, M. (2002). Evidence for diversifying selection in potato virus Y and in the coat protein of other potyviruses. Journal of General Virology, 83, 2563–2573.
Oertel, U., Fuchs, E., & Hohmann, F. (1999). Differentiation of isolates of sugarcane mosaic potyvirus (SCMV) on the basis of molecular, serological and biological investigations. Z PflKrankh Pfl Schutz, 106, 304–313.
Ohshima, K., Tomitaka, Y., Wood Jeffery, T., Minematsu, Y., Kajiyama, H., Tomimura, K., & Gibbs Adrian, J. (2007). Patterns of recombination in turnip mosaic virus genomic sequences indicate hotspots of recombination. Journal of General Virology, 88, 298–315.
Pond, S. L. K., & Frost, S. (2005). Datamonkey: Rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics, 21, 2531–2533.
Simon-Loriere, E., & Holmes, E. C. (2011). Why do RNA viruses recombine? Natural Reviews Microbiology, 9, 617–626.
Stewart, L. R., Bouchard, R., Redinbaugh, M. G., & Meulia, T. (2012). Complete sequence and development of a full-length infectious clone of an Ohio isolate of maize dwarf mosaic virus (MDMV). Virus Research, 165, 219–224.
Tang, W., Yan, Z. Y., Zhu, T. S., Xu, X. J., Li, X. D., & Tian, Y. P. (2018). The complete genomic sequence of Sugarcane mosaic virus from Canna spp. in China. Virology Journal, 15, 147.
Viswanathan, R. (2016). Varietal degeneration in sugarcane and its management in India. Sugar Tech, 18, 1–7.
Viswanathan, R., & Balamuralikrishnan, M. (2005). Impact of mosaic infection on growth and yield of sugarcane. Sugar Tech, 7, 61–65.
Viswanathan, R., & Karuppaiah, R. (2010). Distribution pattern of RNA viruses causing mosaic symptoms and yellow leaf in Indian sugarcane varieties. Sugar Cane International, 28, 202–205.
Viswanathan, R., & Rao, G. P. (2011). Disease scenario and management of major sugarcane diseases in India. Sugar Tech, 13, 336–353.
Viswanathan, R., Balamuralikrishnan, M., & Karuppaiah, R. (2007). Sugarcane mosaic in India: A cause of combined infection of Sugarcane mosaic virus and Sugarcane streak mosaic virus. Sugar Cane International, 25, 6–14.
Viswanathan, R., Balamuralikrishnanan, M., & Karuppaiah, R. (2008). Sugarcane mosaic complex in India: Cause of different viruses/strains. Indian Journal of Virology, 19, 94–94.
Viswanathan, R., Karuppaiah, R., & Balamuralikrishnan, M. (2009). Identification of new variants of SCMV causing sugarcane mosaic in India and assessing their genetic diversity in relation to SCMV type strains. Virus Genes, 39, 375–386.
Viswanathan, R., Karuppaiah, R., & Balamuralikrishnan, M. (2010). Detection of three major RNA viruses infecting sugarcane by multiplex reverse transcription polymerase chain reaction (multiplex RT PCR). Australasian Plant Pathology, 39, 79–84.
Viswanathan, R., Ganesh Kumar, V., Karuppaiah, R., Scindiya, M., & Chinnaraja, C. (2013). Development of duplex immunocapture (duplex IC) RT-PCR for the detection of Sugarcane streak mosaic virus and Sugarcane mosaic virus in sugarcane. Sugar Tech, 15, 399–405.
Wamaitha, M. J., Nigam, D., Maina, S., Stomeo, F., Wangai, A., Njuguna, J. N., Holton, T. A., Wanjala, B. W., Wamalwa, M., Lucas, T., Djikeng, A., & Ruiz, H. G. (2018). Metagenomic analysis of viruses associated with maize lethal necrosis in Kenya. Virology Journal, 15, 90.
Xia, X. C., Melchinger, A. E., Kuntze, L., & Lubberstedt, T. (1999). Quantitative trait loci mapping of resistance to Sugarcane mosaic virus in maize. Phytopathology, 89, 660–667.
Yang, Z. N., & Mirkov, T. E. (1997). Sequence and relationships of sugarcane mosaic and sorghum mosaic virus strains and development of RT-PCR–based RFLPs for strain discrimination. Phytopathology, 87, 932–939.
Zhong, Y., Guo, A., Li, C., Zhuang, B., Lai, M., Wei, C., Luo, J., & Li, Y. (2005). Identification of a naturally occurring recombinant isolate of sugarcane mosaic virus causing maize dwarf mosaic disease. Virus Genes, 30, 75–83.
Acknowledgements
The financial support received from the Department of Biotechnology, New Delhi (BTPR4978-AGR-36712-2012) is greatly acknowledged.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declared that there is no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Bagyalakshmi, K., Parameswari, B. & Viswanathan, R. Phylogenetic analysis and signature of recombination hotspots in sugarcane mosaic virus infecting sugarcane in India. Phytoparasitica 47, 275–291 (2019). https://doi.org/10.1007/s12600-019-00726-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12600-019-00726-1