Abstract
Eukaryotic cells can adapt to endoplasmic reticulum (ER) dysfunction by producing diverse signals from the ER to the cytosol or nucleus. These signaling pathways are collectively known as the unfolded protein response (UPR). The canonical branches of the UPR are mediated by three ER membrane-bound proteins: double-stranded RNA-dependent protein kinase (PKR)-like endoplasmic reticulum kinase (PERK), inositol-requiring enzyme-1 (IRE1) and activating transcription factor 6 (ATF6). These ER stress transducers basically play important roles in cell survival after ER stress. Recently, novel types of ER stress transducers that share a region of high sequence similarity with ATF6 have been identified. They have a transmembrane domain, which allows them to associate with the ER, and possess a transcription-activation domain and a basic leucine zipper (bZIP) domain. These membrane-bound bZIP transcription factors include OASIS, BBF2H7 CREBH, CREB4 and Luman, and are collectively referred to as OASIS family members. Despite their structural similarities with ATF6, differences in activating stimuli and tissue distribution indicate specialized functions of each member on regulating UPR signaling in specific organs and tissues. One of them, OASIS, is expressed preferentially in astrocytes in the central nervous system (CNS). OASIS temporally regulates the differentiation from neural precursor cells into astrocytes to promote the expression of Glial Cell Missing 1 through dynamic interactions among OASIS family members followed by accelerating demethylation of the Gfap promoter. This review is a summary of our current understanding of the physiological functions of OASIS in the CNS.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
ER stress and canonical unfolded protein response
The endoplasmic reticulum (ER) is an essential organelle that regulates the synthesis and post-translational modifications of secretory and membrane proteins. Nascent proteins are folded in the ER with the assistance of molecular chaperones and folding enzymes. Various pathophysiological conditions, such as ER-calcium depletion, oxidative stress, hypoglycemia, expression of mutated proteins and hypoxia, interfere with the correct folding of proteins followed by promoting the accumulation of these unfolded or misfolded proteins in the ER lumen. These conditions are collectively termed ER stress, and have the potential to induce cellular damage. The ER responds to these perturbations by activating an integrated signal transduction pathway, called the unfolded protein response (UPR) (Ron 2002; Kaufman 2002; Schroder and Kaufman 2005). Activation of the UPR leads to a transient translational inhibition, followed by the activation of transcription of ER molecular chaperones and the degradation of unfolded proteins accumulated in the ER [ER-associated degradation (ERAD)]. In mammalian cells, ER stress-sensing and UPR signaling are regulated by three well-established ER stress transducers: double-stranded RNA-dependent protein kinase (PKR)-like endoplasmic reticulum kinase (PERK) (Harding et al. 1999), inositol-requiring enzyme-1 (IRE1) (Tirasophon et al. 1998; Urano et al. 2000), and activating transcription factor 6 (ATF6) (Li et al. 2000; Yoshida et al. 2000). PERK directly phosphorylates the α subunit of eukaryotic initiation factor (eIF2α) and leads to shut-down of most cellular protein synthesis (Harding et al. 2000). IRE1, ATF6 and ATF4, which is downstream of PERK-eIF2α pathway, are involved in transcriptional regulation of ER molecular chaperones and ERAD-related genes. If these strategies fail, cells undergo ER stress-induced apoptosis (Ron 2002; Kaufman 2002; Schroder and Kaufman 2005). Thus, cell fates after ER stress are determined by the balance of cell survival and death signals regulated by the UPR (Fig. 1).
Schema of endoplasmic reticulum (ER) stress and unfolded protein response (UPR) signaling. Activation of the UPR leads to a transient translational inhibition, followed by the activation of transcription of ER molecular chaperones and the degradation of the unfolded proteins accumulated in the ER [ER-associated degradation (ERAD)]. If these strategies fail, cells undergo ER stress-induced apoptosis. Ask1 apoptosis signal-regulating kinase 1, ATF4 activating transcription factor 4, CHOP C/EBP homologous protein, JNK c-jun N-terminal kinase, RIP regulated intramembrane proteolysis, TRAF2 TNF receptor-associated factor 2, XBP1 X-box binding protein 1, P phosphorylation
Novel types of membrane-bound basic leucine zipper transcription factors activated by regulated intramembrane proteolysis
In humans, there are more than 55 known basic leucine zipper (bZIP) transcription factors (Newman and Keating 2003). By sequence similarity in the coiled-coil region, these transcription factors can be divided into 16 different families. Recently, novel types of ER stress transducers, i.e., ER membrane-bound bZIP transcription factors that share a region of high sequence similarity with ATF6, have been identified. They have a transmembrane domain that allows them to associate with the ER, and possess both a transcription-activation domain and a bZIP domain. The new types of ER stress transducers include OASIS (Kondo et al. 2005; Murakami et al. 2009), BBF2H7 (Kondo et al. 2007; Saito et al. 2009), CREBH (Omori et al. 2001; Zhang et al. 2006), CREB4 (Qi et al. 2002; Cao et al. 2002; Adham et al. 2005; Nagamori et al. 2005), and Luman (Lu et al. 1997; DenBoer et al. 2005; Liang et al. 2006) (Fig. 2). Interestingly, each of them commonly contains the consensus sequence for cleavage by site-1 protease (S1P) and site-2 protease (S2P) in its luminal segment, indicating that they are processed at transmembrane regions by regulated intramembrane proteolysis (RIP) (Ye et al. 2000; Bailey and O’Hare 2007). Their cleaved N-terminal fragments translocate into the nucleus to act as transcription factors. In this review, these bZIP transmembrane transcription factors are referred to as OASIS family members. Despite structural similarities among these proteins and ATF6, differences in activating stimuli and tissue distribution indicate that these proteins play specific roles in regulating the UPR signaling in specific organs and tissues. OASIS, BBF2H7, CREBH, and CREB4 are expressed preferentially in osteoblasts and astrocytes (Kondo et al. 2005; Murakami et al. 2009), chondrocytes (Saito et al. 2009), liver cells (Omori et al. 2001), and prostate and testis (Cao et al. 2002), respectively. The Luman transcript is present in a wide range of adult and fetal tissues (Lu et al. 1997), but its translated product has been found only in trigeminal ganglional neurons and monocytes, and dendritic cells (DCs) (Lu and Misra 2000; Ko et al. 2004; Eleveld-Trancikova et al. 2010). Evolutionarily, orthologues of ATF6 and OASIS family proteins exist in organisms higher than Caenorhabditis elegans. C. elegans contains three bZIP transcription factors. Although the degree of homology is less than those in the mammalian family, these proteins are related most closely to ATF6, OASIS and Luman (Shen et al. 2005; Fox et al. 2010). In Drosophila, two bZIP transcription factors are related most closely to ATF6 and OASIS (Fox et al. 2010). In vertebrates, cells are diversely differentiated to play proper roles for various biological phenomena, and also adapt to environmental parameters. Vertebrate cells adjust the functionality and capacity of their ER depending on the diversity of cell types. Thus, the signaling of the vertebrate UPR, which is regulated by canonical ER stress transducers and OASIS family members, has considerable complexity and is highly developed in cell-type specific patterns. This review is focused on the current understanding of the biological characteristics and physiological functions of OASIS in the central nervous system (CNS).
Predicted peptide features of mouse OASIS, BBF2H7, CREBH, CREB4, Luman and ATF6. Novel types of ER stress transducers, OASIS family members including OASIS, BBF2H7, CREBH, CREB4 and Luman share a region of high sequence similarity with ATF6. They have a transmembrane domain, which allows them to associate with the ER, and possess a transcription-activation domain and a bZIP domain
Structure and activation mechanisms of OASIS
The Oasis gene was originally identified as a gene specifically induced in long-term cultured astrocytes (Honma et al. 1999). The OASIS protein is a bZIP transcription factor of the cyclic AMP-response element (CRE)-binding protein (CREB)/ATF family, with a transmembrane domain that allows it to associate with the ER. Its N-terminus containing the transmembrane domain is 31 % identical to ATF6, but its C-terminus, which is within the ER lumen, does not show homology to ATF6. In the luminal segment, OASIS contains the sequence RSLL (beginning at residue 423), which fits the RxxL consensus for S1P whose active site faces the Golgi lumen. Indeed, under ER stress conditions, OASIS is cleaved at the site by S1P and subsequently at the transmembrane domain by S2P (Kondo et al. 2005; Murakami et al. 2006). The cleaved N-terminus containing transcription-activation domain and bZIP domain translocates into the nucleus to act as a transcription factor through the binding to the CRE sequence (Fig. 3). ATF6 contains a stretch of amino acid sequence in its luminal domain that is essential for translocation from the ER to the Golgi apparatus (Shen et al. 2002). However, translocation to the Golgi apparatus and proteolytic processing are intact in all deletion mutants for the luminal domain of OASIS, indicating that OASIS does not have significant sequences for Golgi localization signaling (Murakami et al. 2006). Therefore, the translocation system from the ER to the Golgi apparatus is quite different between OASIS and ATF6 under ER stress conditions. Under normal conditions, ATF6 is bound constitutively by an ER-resident chaperone BiP in its luminal domain and rendered inactive (Shen et al. 2002; Chen et al. 2002). Accumulation of unfolded proteins in the ER results in the dissociation of BiP from the luminal domain of ATF6 and then ATF6 is translocated to the Golgi apparatus. The ER luminal domain of OASIS does not possess the sensing function for unfolded proteins. OASIS is degraded easily under normal conditions via the ubiquitin–proteasome pathway (Kondo et al. 2012). An ER stress condition enhances the stability of OASIS. The stabilized OASIS is transported automatically from the ER to the Golgi apparatus and is then cleaved to generate N-terminus by RIP. Thus, the ER stress-sensing function is not necessary in the ER luminal domain of OASIS for its activation.
Physiological functions of OASIS in skeletal systems
Oasis is highly expressed in osteoblasts of osseous tissues (Murakami et al. 2009) and astrocytes of the CNS (Kondo et al. 2005; Chihara et al. 2009), and to considerable levels in intestine, salivary glands, and prostate (Honma et al. 1999; Omori et al. 2002; Asada et al. 2012). In C6 glioma cell lines and primary cultured osteoblasts, Oasis is induced at the transcriptional level after treatment with ER stressors such as tunicamycin or thapsigargin (Kondo et al. 2005). Oasis deficient (Oasis −/−) mice are born at the expected Mendelian ratios, but the mice exhibit severe osteopenia involving a decrease in bone density at all skeletal sites and spontaneous fractures (Murakami et al. 2009). From detailed analyses on gene expression in Oasis −/− bones, Type I collagen (Col1)—a major component of osseous tissues—was identified as one of the targets for OASIS. OASIS activates the transcription of Col1 through direct binding to a CRE-like sequence that exists in the Col1 promoter regions. Indeed, Oasis −/− osseous tissues show a significant decrease in the amounts of type I collagen. The defect of bone formation involving decreased amounts of type I collagen is rescued completely by osteoblast-specific overexpression of OASIS, indicating that osteopenia in Oasis −/− mice is caused primarily by deletion of Oasis and its target, Col1 gene in osteoblasts (Murakami et al. 2010).
Oasis −/− mice exhibit decreased numbers of astrocytes
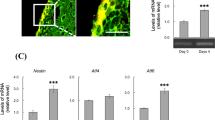
As mentioned above, Oasis is also highly expressed in astrocytes of the CNS. OASIS is up-regulated in reactive astrocytes after neuronal degeneration induced by kainic acid (KA). In hippocampal astrocytes injured by intraperitoneal injection of KA, ER stress is induced and Oasis mRNA is strongly up-regulated (Chihara et al. 2009). Pyramidal neurons in the hippocampi of Oasis −/− mice are more susceptible to the toxicity induced by KA than those of wild-type (WT) mice. The number of glial fibrillary acidic protein (GFAP)-positive reactive astrocytes, an astrocyte marker, was decreased in the hippocampi of Oasis −/− mice compared with those of WT mice after brain injury. In embryonic stages, Oasis mRNA is barely observed in the cerebral cortices of embryonic day (E) 14.5 mice; however, strong signals are detected at E16.5 and E18.5 (Saito et al. 2012), coinciding with the initiation of extensive differentiation of neural precursor cells (NPCs) into astrocytes. Oasis −/− mice exhibit impaired astrocyte differentiation in embryonic stages. The numbers of cells positive for GFAP are significantly higher in the cerebral cortices of WT mice than in those of Oasis −/− mice at E18.5 (Fig. 4a, b) (Saito et al. 2012). By contrast, those of nestin—an NPC marker—are higher in the cerebral cortices of Oasis −/− mice than in those of WT mice at E18.5 (Fig. 4c, d). The impaired astrocyte differentiation is also observed in primary cultured NPCs prepared from E14.5 Oasis −/− mice telencephalons. In primary cultured Oasis −/− NPCs treated with leukemia inhibitory factor (LIF) and bone morphogenetic protein 2 (BMP2), which promote the differentiation of NPCs into astrocytes (Bonni et al. 1997; Nakashima et al. 1999a, b) (Fig. 4e), the induction of Gfap expression and the reduction of Nestin expression are significantly inhibited compared with those of WT cells (Fig. 4f).
Differentiation of neural precursor cells (NPCs) into astrocytes is delayed in Oasis −/− mice. Immunohistochemical analysis of glial fibrillary acidic protein (GFAP) (a, b) and nestin (c, d) in the cerebral cortices of E18.5 WT and Oasis −/− mice. The number of GFAP-positive cells was lower, and that of nestin-positive cells was higher in Oasis −/− mice. Yellow lines Surfaces of cerebral cortices. Bar 100 μM. e Primary cultured NPCs were prepared from the telencephalons of E14.5 WT and Oasis −/− mice. Cells were treated with leukemia inhibitory factor (LIF) and bone morphogenetic protein 2 (BMP2) to promote the differentiation of NPCs into astrocytes. f RT-PCR analysis of Gfap and Nestin in primary cultured NPCs treated with LIF and BMP2 for the indicated times. The up-regulation of Gfap and the down-regulation of Nestin are inhibited in Oasis −/− cells
The target of OASIS in NPCs is Gcm1
Glial Cell Missing 1 (Gcm1) is identified as a target of OASIS. OASIS promotes the expression of Gcm1 through the binding to the CRE-like sequence in the promoter region of Gcm1 in CNS and trophoblasts (Schubert et al. 2008; Saito et al. 2012). The Gcm1 Drosophila ortholog, gcm, has been reported to control neuronal and glial fates (Hosoya et al. 1995; Jones et al. 1995; Vincent et al. 1996). In mammalian cells, the introduction of Gcm1 into cultured brain cells derived from mice induced the astrocyte lineage (Iwasaki et al. 2003). As described above, Gcm1 is critical for astrocyte differentiation; however, the detailed mechanisms regulating differentiation have not been defined. It is well known that the activation of Stat3 and Smads, and epigenetic regulation of the methylation status of the Gfap promoter, are essential for the differentiation of NPCs into astrocytes. Phosphorylated-Stat3 and Smads bind to demethylated sites in the Gfap promoter to form a Stat3-Smads-p300 complex and promote transcription of Gfap (Bonni et al. 1997; Nakashima et al. 1999a, b, Takizawa et al. 2001). Although the levels of phosphorylated Stat3 and Smads are not changed, the amount of demethylation of the Gfap promoter is decreased significantly in Oasis −/− NPCs compared with those of WT NPCs (Saito et al. 2012). Thus, the delayed astrocyte differentiation in Oasis −/− cells is caused by inhibition of demethylation of the Gfap promoter (Fig. 5). The mammalian homologs of gcm are Gcm1 and Gcm2. These molecules show little homology and have no common functional domains except for the GCM-motif (Akiyama et al. 1996). Previous report have shown that both have the potential to accelerate demethylation of the Hes5 promoter by direct binding to the GCM-binding site in the promoter followed by acquiring stem cell properties, but Gcm2 is a more crucial factor than Gcm1 for demethylation of the Hes5 promoter because of the lower expression of Hes5 in Gcm2 −/− mice than in Gcm1 −/− mice (Hitoshi et al. 2011). The expression of Gcm1 is significantly up-regulated at later stages of mouse embryonic development, whereas that of Gcm2 is transiently up-regulated at the early stage (Hitoshi et al. 2011). Therefore, we presume that Gcm2 is involved mainly in demethylation of the Hes5 promoter at the early stage of mouse embryonic development and, conversely, that Gcm1 mainly plays a role in the demethylation of the Gfap promoter in the late stage. The distinct expression patterns and target promoters for demethylation between Gcm1 and Gcm2 may determine glial and neuronal cell lineages, respectively.
OASIS-Gcm1 signaling is involved in demethylation of the Gfap promoter. Activation of Stat3 and Smads, and epigenetic regulation of the methylation status of the Gfap promoter, are essential for differentiation of NPCs into astrocytes. Stat3 and Smads are phosphorylated downstream of LIF and BMP2, and phosphorylated Stat3 and Smads bind to demethylated sites in the Gfap promoter to promote transcription of Gfap. OASIS-Gcm1 signaling does not alter the levels of phosphorylated Stat3 and Smads, but regulates the demethylation status of the Gfap promoter region
Gcm1 expression modulation by OASIS family members
The bZIP type transcription factors including OASIS family members are known to form homodimers or heterodimers to promote transcription (Hai and Hartman 2001; Zhang et al. 2006). During astrocyte differentiation, not only OASIS but also CREB4 and Luman regulate the transcription of Gcm1 (Saito et al. 2012). OASIS and CREB4 N-termini form heterodimers and activate transcription of Gcm1 during astrocyte differentiation. Conversely, the Luman N-terminus inhibits the formation of the OASIS-CREB4 heterodimer by binding to the OASIS N-terminus and down-regulating transcription of Gcm1, leading to a cessation of astrocyte differentiation (Fig. 6). Therefore, dynamic interactions among OASIS, CREB4 and Luman regulate the spatio-temporal expression of Gcm1 during astrocyte differentiation. These OASIS family members activate UPR signaling in response to physiological ER stress to fine-tune the expression of Gcm1 followed by regulating astrocyte differentiation. UPR signaling activated by physiological ER stress is known to be crucial for the differentiation of secretory cells including osteoblasts (Murakami et al. 2009; Saito et al. 2011; Tohmonda et al. 2011), chondrocytes (Saito et al. 2009), and plasma cells (Gass et al. 2002; Iwakoshi et al. 2003). During the differentiation of progenitor cells into these mature secretory cells, secretory materials are gradually produced, and abundant nascent proteins are delivered to the ER. Such an event may serve as a trigger for physiological ER stress. Astrocytes also synthesize and secrete various neurotrophic factors and cytokines. Physiological ER stress caused by production of abundant secretory proteins could be activated during differentiation of NPCs into mature astrocytes. OASIS, CREB4 and Luman may be activated as transcription factors in response to this physiological ER stress. The difference between physiological and pathological ER stress remains unclear. Treatment of NPCs with low doses of several ER stressors, including tunicamycin and dithiothreitol, promotes the differentiation of NPCs into astrocytes but not apoptosis (Saito et al. 2012). Thus, physiological and pathological ER stress may be distinguished, in part, only by the difference of the burden levels on the ER. Additionally, recent studies have shown that ER stress transducers can mildly activate ER stress-independent phenomena such as changes in the lipid-constitution of the ER membrane (Volmer et al. 2013). It is possible that not only mild ER stress, which does not promote apoptosis, but also ER stress-independent phenomena may activate ER stress transducers in the same was as physiological ER stress during cell differentiation.
OASIS, CREB4 and Luman N-termini form heterodimers to regulate Gcm1 expression. OASIS-CREB4 heterodimers activate transcription of Gcm1 during astrocyte differentiation. Conversely, the Luman N-terminus inhibits the formation of OASIS-CREB4 heterodimers by binding to the OASIS N-terminus, down-regulating transcription of Gcm1
Conclusion
UPR signaling was originally found as a system for evading cellular damage in acute ER dysfunction. However, recent advanced studies have revealed that UPR signaling provides important signals for regulating cellular physiology. Not only canonical ER stress transducers, but also OASIS family members act as signaling centers for numerous networks originating from the ER. In particular, OASIS family members are specialized in biological regulation, including cell differentiation, maturation and maintenance of basal cellular homeostasis. Thus far, ER stress damages cells, and has been seen as a major cause of various diseases (Schroder and Kaufman 2005; Hotamisligil 2010). However, recent findings, as shown here, suggest that an appropriate ER stress provides an important signal for controlling cellular physiology. It would not be surprising that differentiation, maturation and functional regulations of other cells are similarly regulated by physiological ER stress. OASIS is involved in the differentiation of osteoblasts and goblet cells in the large intestine as well as in astrocytes (Murakami et al. 2009; Asada et al. 2012; Saito et al. 2012) (Fig. 7). Further, two previous reports regarding Creb4 −/− mice revealed that this molecule plays a role in differentiation of sperm cells (Adham et al. 2005; Nagamori et al. 2006). BBF2H7 and Luman are involved in the differentiation of chondrocytes and DCs, respectively (Saito et al. 2009; Eleveld-Trancikova et al. 2010). CREBH, another OASIS family member, plays a role in activating expression of inflammatory response in the liver (Zhang et al. 2006). Besides OASIS family members, canonical ER stress transducers have also been defined to play roles in development, differentiation, and maturation of cells. IRE1-XBP-1 signaling is particularly important in cells that are active in protein secretion, such as antibody-secretory plasma cells, hepatocytes, and exocrine pancreatic cells (Calfon et al. 2002; Gass et al. 2002; Iwakoshi et al. 2003; Lee et al. 2005). PERK-eIF2α signaling mediates differentiation in insulin-secreting beta-cells and osteoblasts (Harding et al. 2001; Zhang et al. 2002; Wei et al. 2008; Saito et al. 2011). Thus, functional regulation mediated by physiological UPR signaling has been demonstrated to associate with various biological phenomena. Further studies including analyses on the detailed signaling and functions of the UPR components including OASIS family members are needed to better understand the diversity of the UPR system in mammals.
OASIS activated by physiological ER stress promotes various cell differentiation and maturation. Differentiation stimuli to undifferentiated cells initiates differentiation. Altered ER environments, such as the synthesis of abundant secretory proteins, causes physiological ER stress. Physiological ER stress activates OASIS followed by accelerating the differentiation and maturation of cells including astrocytes, osteoblasts and goblet cells
The signaling from the ER mediated by OASIS family members is presumed to be triggered by ER perturbation. However, is ER stress actually only the stimulation for activating OASIS family members that are required for initiating physiologically important signaling events? Indeed, OASIS is weakly expressed in hippocampal neurons under control conditions, although it is not activated by KA-induced ER stress (Chihara et al. 2009). These facts suggest that OASIS activation may be regulated by other cell-specific mechanisms as well as ER stress. Recently, it has been reported that ER stress transducers can be activated in response to ER stress-independent phenomena such as the changes in lipid-constitution of the ER membrane (Volmer et al. 2013). Further, OASIS does not have the ER stress-sensing domain in its ER luminal domain for activation. These facts indicate the possibility that OASIS family members, including OASIS, may be also activated by other mechanisms or signals including cell-specific differentiating stimuli, which is different from ER stress. Elucidation of the mechanisms responsible for their activation in specific tissues and cells may lead to clarification of the physiological functions of these OASIS family members. In addition, it is widely known that bZIP type transcription factors can form homodimers or heterodimers, and that this dimerization often affects their binding ability. As shown in this review, the OASIS N-terminus binds to CREB4 and Luman N-termini and forms heterodimers to fine-tune the expression of Gcm1 in the CNS (Saito et al. 2012). ATF6 can bind to active forms of CREBH and serve as a potent enhancer to augment CREBH-mediated acute phase response gene transcription for the acute inflammatory response (Zhang et al. 2006). It is possible that changing the binding partners for each transcription factor may switch the target genes promoting expression, thus allowing fine-tuning of UPR signaling for regulating physiological phenomena. OASIS family members reveal unique cell- or tissue-specific expression patterns, but overlapping expression is also observed in each protein in the same cells or tissues. Thus, it is necessary to analyze the spatio-temporal hetero- or homo-dimerization of each protein in vivo to better understand the diversity of the UPR system in mammals.
References
Adham IM, Eck TJ, Mierau K, Müller N, Sallam MA, Paprotta I, Schubert S, Hoyer-Fender S, Engel W (2005) Reduction of spermatogenesis but not fertility in Creb3l4-deficient mice. Mol Cell Biol 25:7657–7664
Akiyama Y, Hosoya T, Poole AM, Hotta Y (1996) The gcm-motif: a novel DNA-binding motif conserved in Drosophila and mammals. Proc Natl Acad Sci USA 93:14912–14916
Asada R, Saito A, Kawasaki N, Kanemoto S, Iwamoto H, Oki M, Miyagi H, Izumi S, Imaizumi K (2012) The endoplasmic reticulum stress transducer OASIS is involved in the terminal differentiation of goblet cells in the large intestine. J Biol Chem 287:8144–8153
Bailey D, O’Hare P (2007) Transmembrane bZIP transcription factors in ER stress signaling and the unfolded protein response. Antioxid Redox Signal 9:2305–2321
Bonni A, Sun Y, Nadal-Vicens M, Bhatt A, Frank DA, Rozovsky I, Stahl N, Yancopoulos GD, Greenberg ME (1997) Regulation of gliagenesis in the central nervous system by JAK-STAT signaling pathway. Science 278:477–483
Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, Clark SG, Ron D (2002) IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature 415:92–96
Cao G, Ni X, Jiang M, Ma Y, Cheng H, Guo L, Ji C, Gu S, Xie Y, Mao Y (2002) Molecular cloning and characterization of a novel human cAMP response element-binding (CREB) gene (CREB4). J Hum Genet 47:373–376
Chen X, Shen J, Prywes R (2002) The luminal domain of ATF6 senses endoplasmic reticulum (ER) stress and causes translocation of ATF6 from the ER to the Golgi. J Biol Chem 277:13045–13052
Chihara K, Saito A, Murakami T, Hino S, Aoki Y, Sekiya H, Aikawa Y, Wanaka A, Imaizumi K (2009) Increased vulnerability of hippocampal pyramidal neurons to the toxicity of kainic acid in OASIS-deficient mice. J Neurochem 110:956–965
DenBoer LM, Hardy-Smith PW, Hogan MR, Cockram GP, Audas TE, Lu R (2005) Luman is capable of binding and activating transcription from the unfolded protein response element. Biochem Biophys Res Commun 331:113–119
Eleveld-Trancikova D, Sanecka A, van Hout-Kuijer MA, Looman MVG, Hendriks IAM, Jansen BJH, Adema GJ (2010) DC-STAMP interacts with ER-resident transcription factor LUMAN which becomes activated during DC maturation. Mol Immunol 47:1963–1973
Fox RM, Hanlon CD, Andrew DJ (2010) The CrebA/Creb3-like transcription factors are major and direct regulators of secretory capacity. J Cell Biol 191:479–492
Gass JN, Gifford NM, Brewer JW (2002) Activation of an unfolded protein response during differentiation of antibody-secreting B cells. J Biol Chem 277:49047–49054
Hai T, Hartman MG (2001) The molecular biology and nomenclature of the activating transcription factor/cAMP responsive element binding family of transcription factors: activating transcription factor proteins and homeostasis. Gene 273:1–11
Harding HP, Zhang Y, Ron D (1999) Protein translation and folding are coupled by an endoplasmic-reticulum-resident kinase. Nature 397:271–274
Harding HP, Novoa I, Zhang Y, Zeng H, Wek R, Schapira M, Ron D (2000) Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol Cell 6:1099–1108
Harding HP, Zeng H, Zhang Y, Jungries R, Chung P, Plesken H, Sabatini DD, Ron D (2001) Diabetes mellitus and exocrine pancreatic dysfunction in perk−/− mice reveals a role for translational control in secretory cell survival. Mol Cell 7:1153–1163
Hitoshi S, Ishino Y, Kumar A, Jasmine S, Tanaka KF, Kondo T, Kato S, Hosoya T, Hotta Y, Ikenaka K (2011) Mammalian Gcm genes induce Hes5 expression by active DNA demethylation and induce neural stem cells. Nat Neurosci 14:957–964
Honma Y, Kanazawa K, Mori T, Tanno Y, Tojo M, Kiyosawa H, Takeda J, Nikaido T, Tsukamoto T, Yokoya S, Wanaka A (1999) Identification of a novel gene, OASIS, which encodes for a putative CREB/ATF family transcription factor in the long-term cultured astrocytes and gliotic tissue. Brain Res Mol Brain Res 69:93–103
Hosoya T, Takizawa K, Nitta K, Hotta Y (1995) Glial cells missing: a binary switch between neuronal and glial determination in Drosophila. Cell 82:1025–1036
Hotamisligil GS (2010) Endoplasmic reticulum stress and the inflammatory basis of metabolic disease. Cell 140:900–917
Iwakoshi NN, Lee AH, Vallabhajosyula P, Otipoby KL, Rajewsky K, Glimcher LH (2003) Plasma cell differentiation and the unfolded protein response intersect at the transcription factor XBP-1. Nat Immunol 4:321–329
Iwasaki Y, Hosoya T, Takebayashi H, Ogawa Y, Hotta Y, Ikenaka K (2003) The potential to induce glial differentiation is conserved between Drosophila and mammalian glial cells missing genes. Development 130:6027–6035
Jones BW, Fetter RD, Tear G, Goodman CS (1995) Glial cells missing: a genetic switch that controls glial versus neuronal fate. Cell 82:1013–1023
Kaufman RJ (2002) Orchestrating the unfolded protein response in health and disease. J Clin Invest 110:1389–1398
Ko J, Jang SW, Kim YS, Kim IS, Sung HJ, Kim HH, Park JY, Lee YH, Kim J, Na DS (2004) Human LZIP binds to CCR1 and differentially affects the chemotactic activities of CCR1-dependent chemokines. FASEB J 18:890–892
Kondo S, Murakami T, Tatsumi K, Ogata M, Kanemoto S, Otori K, Iseki K, Wanaka A, Imaizumi K (2005) OASIS, a CREB/ATF-family member, modulates UPR signalling in astrocytes. Nat Cell Biol 7:186–194
Kondo S, Saito A, Hino S-I, Murakami T, Ogata M, Kanemoto S, Nara S, Yamashita A, Yoshinaga K, Hara H, Imaizumi K (2007) BBF2H7, a novel transmembrane bZIP transcription factor, is a new type of endoplasmic reticulum stress transducer. Mol Cell Biol 27:1716–1729
Kondo S, Hino S-I, Saito A, Kanemoto S, Kawasaki N, Asada R, Izumi S, Iwamoto H, Oki M, Miyagi H, Kaneko M, Nomura Y, Urano F, Imaizumi K (2012) Activation of OASIS family, ER stress transducers, is dependent on its stabilization. Cell Death Differ 19:1939–1949
Lee AH, Chu GC, Iwakoshi NN, Glimcher LH (2005) XBP-1 is required for biogenesis of cellular secretory machinery of exocrine glands. EMBO J 24:4368–4380
Li M, Baumeister P, Roy B, Phan T, Foti D, Luo S, Lee AS (2000) ATF6 as a transcription activator of the endoplasmic reticulum stress element: thapsigargin stress-induced changes and synergistic interactions with NF-Y and YY1. Mol Cell Biol 20:5096–5106
Liang G, Audas TE, Li Y, Cockram GP, Dean JD, Martyn AC, Kokame K, Lu R (2006) Luman/CREB3 induces transcription of the endoplasmic reticulum (ER) stress response protein Herp through an ER stress response element. Mol Cell Biol 26:7999–8010
Lu R, Misra V (2000) Potential role for luman, the cellular homologue of herpes simplex virus VP16 (alpha gene trans-inducing factor), in herpesvirus latency. J Virol 74:934–943
Lu R, Yang P, O’Hare P, Misra V (1997) Luman, a new member of the CREB/ATF family, binds to herpes simplex virus VP16-associated host cellular factor. Mol Cell Biol 17:5117–5126
Murakami T, Kondo S, Ogata M, Kanemoto S, Saito A, Imaizumi K (2006) Cleavage of the membrane-bound transcription factor OASIS in response to endoplasmic reticulum stress. J Neurochem 96:1090–1100
Murakami T, Saito A, Hino S-I, Kondo S, Kanemoto S, Chihara K, Sekiya H, Tsumagari K, Ochiai K, Yoshinaga K, Saitoh M, Nishimura R, Yoneda T, Kou I, Furuichi T, Ikegawa S, Ikawa M, Okabe M, Wanaka A, Imaizumi K (2009) Signalling mediated by the endoplasmic reticulum stress transducer OASIS is involved in bone formation. Nat Cell Biol 11:1205–1211
Murakami T, Hino S-I, Nishimura R, Yoneda T, Wanaka A, Imaizumi K (2010) Distinct mechanisms are responsible for osteopenia and growth retardation in OASIS-deficient mice. Bone 48:514–523
Nagamori I, Yabuta N, Fujii T, Tanaka H, Yomogida K, Nishimune Y, Nojima H (2005) Tisp40, a spermatid specific bZip transcription factor, functions by binding to the unfolded protein response element via the Rip pathway. Genes Cells 10:575–594
Nagamori I, Yomogida K, Ikawa M, Okabe M, Yabuta N, Nojima H (2006) The testes-specific bZip type transcription factor Tisp40 plays a role in ER stress responses and chromatin packaging during spermiogenesis. Genes Cells 11:1161–1171
Nakashima K, Yanagisawa M, Arakawa H, Kimura N, Hisatsune T, Kawabata M, Miyazono K, Taga T (1999a) Synergistic signaling in fetal brain by STAT3-Smad1 complex bridged by p300. Science 284:479–482
Nakashima K, Wiese S, Yanagisawa M, Arakawa H, Kimura N, Hisatsune T, Yoshida K, Kishimoto T, Sendtner M, Taga T (1999b) Developmental requirement of gp130 signaling in neuronal survival and astrocyte differentiation. J Neurosci 19:5429–5434
Newman JR, Keating AE (2003) Comprehensive identification of human bZIP interactions with coiled-coil arrays. Science 300:2097–2101
Omori Y, Imai J, Watanabe M, Komatsu T, Suzuki Y, Kataoka K, Watanabe S, Tanigami A, Sugano S (2001) CREB-H: a novel mammalian transcription factor belonging to the CREB/ATF family and functioning via the box-B element with a liver-specific expression. Nucleic Acids Res 29:2154–2162
Omori Y, Imai J, Suzuki Y, Watanabe S, Tanigami A, Sugano S (2002) OASIS is a transcriptional activator of CREB/ATF family with a transmembrane domain. Biochem Biophys Res Commun 293:470–477
Qi H, Fillion C, Labrie Y, Grenier J, Fournier A, Berger L, El-Alfy M, Labrie C (2002) AIbZIP, a novel bZIP gene located on chromosome 1q21.3 that is highly expressed in prostate tumors and of which the expression is up-regulated by androgens in LNCaP human prostate cancer cells. Cancer Res 62:721–733
Ron D (2002) Translational control in the endoplasmic reticulum stress response. J Clin Invest 110:1383–1388
Saito A, Hino S-I, Murakami T, Kanemoto S, Kondo S, Saitoh M, Nishimura R, Yoneda T, Furuichi T, Ikegawa S, Ikawa M, Okabe M, Imaizumi K (2009) Regulation of endoplasmic reticulum stress response by a BBF2H7-mediated Sec23a pathway is essential for chondrogenesis. Nat Cell Biol 11:1197–1204
Saito A, Ochiai K, Kondo S, Tsumagari K, Murakami T, Cavener DR, Imaizumi K (2011) ER stress response mediated by the perk-eIF2α-ATF4 pathway is involved in osteoblast differentiation induced by BMP2. J Biol Chem 286:4809–4818
Saito A, Kanemoto S, Kawasaki N, Asada R, Iwamoto H, Oki M, Miyagi H, Izumi S, Sanosaka T, Nakashima K, Imaizumi K (2012) Unfolded protein response activated by OASIS family transcription factors, promotes astrocyte differentiation. Nat Commun 3:967. doi:10.1038/ncomms1971
Schroder M, Kaufman RJ (2005) ER stress and the unfolded protein response. Mut Res 569:29–63
Schubert SW, Abendroth A, Kilian K, Vogler T, Mayr B, Knerr I, Hashemolhosseini S (2008) bZIP-Type transcription factors CREB and OASIS bind and stimulate the promoter of the mammalian transcription factor GCMa/Gcm1 in trophoblast cells. Nucleic Acids Res 36:3834–3846
Shen J, Chen X, Hendershot L, Prywes R (2002) ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev Cell 3:99–111
Shen X, Ellis RE, Sakaki K, Kaufman RJ (2005) Genetic interactions due to constitutive and inducible gene regulation mediated by the unfolded protein response in C. elegans. PLoS Genet 1:355–368
Takizawa T, Nakashima K, Namihira M, Ochiai W, Uemura A, Yanagisawa M, Fujita N, Nakao M, Taga T (2001) DNA methylation is a critical cell-intrinsic determinant of astrocyte differentiation in the fetal brain. Dev Cell 1:749–758
Tirasophon W, Welihinda AA, Kaufman RJ (1998) A stress response pathway from the endoplasmic reticulum to the nucleus requires a novel bifunctional protein kinase/endoribonuclease (Ire1p) in mammalian cells. Genes Dev 12:1812–1824
Tohmonda T, Miyauchi Y, Ghosh R, Yoda M, Uchikawa S, Takito J, Morioka H, Nakamura M, Iwawaki T, Chiba K, Toyama Y, Urano F, Horiuchi K (2011) The IRE1α-XBP1 pathway is essential for osteoblasts differentiation through promoting transcription of Osterix. EMBO Rep 12:451–457
Urano F, Wang X, Bertolotti A, Zhang Y, Chung P, Harding HP, Ron D (2000) Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science 287:664–666
Vincent S, Vonesch JL, Giangrande A (1996) Glide directs glial fate commitment and cell fate switch between neurones and glia. Development 122:131–139
Volmer R, van der Ploeg K, Ron D (2013) Membrane lipid saturation activates endoplasmic reticulum unfolded protein response transducers through their transmembrane domains. Proc Natl Acad Sci USA 110:4628–4633
Wei J, Sheng X, Feng D, McGrath B, Cavener DR (2008) PERK is essential for neonatal skeletal development to regulate osteoblast proliferation and differentiation. J Cell Physiol 217:693–707
Ye J, Rawson RB, Komuro R, Chen X, Davé UP, Prywes R, Brown MS, Goldstein JL (2000) ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol Cell 6:1355–1364
Yoshida H, Okada T, Haze K, Yanagi H, Yura T, Negishi M, Mori K (2000) ATF6 activated by proteolysis binds in the presence of NF-Y (CBF) directly to the cis-acting element responsible for the mammalian unfolded protein response. Mol Cell Biol 20:6755–6767
Zhang P, McGrath B, Li S, Frank A, Zambito F, Reinert J, Gannon M, Ma K, McNaughton K, Cavener DR (2002) The PERK eukaryotic initiation factor 2 alpha kinase is required for the development of the skeletal system, postnatal growth, and the function and viability of the pancreas. Mol Cell Biol 22:3864–3874
Zhang K, Shen X, Wu J, Sakaki K, Saunders T, Rutkowski DT, Back SH, Kaufman RJ (2006) Endoplasmic reticulum stress activates cleavage of CREBH to induce a systemic inflammatory response. Cell 124:587–599
Acknowledgments
The author thanks M. Tohyama, S. Shiosaka, K. Yoshinaga and S-I. Hino for helpful discussions. The author also thanks the members of our laboratory, K. Imaizumi, S. Kanemoto, Y. Zhang, M. Kamon, R. Asada, K. Hino, Y. Tatsumoto, H. Iwamoto and H. Miyagi for valuable discussions and experimental support related to this work. K. Nakashima and T. Sanosaka provided substantial input into the experiments of astrocyte differentiation. This work was partly supported by grants from the Japan Society for the Promotion of Science KAKENHI (#22020030, #22800049), Sumitomo Foundation, Mochida Memorial Foundation for Medical and Pharmaceutical Research, Astellas Foundation for Research on Metabolic Disorders, Takeda Science Foundation, The Pharmacological Research Foundation Tokyo, Daiichi-Sankyo Foundation of Life Science, The Naito Foundation, Senri Life Science Foundation, Hokuto Foundation for Bioscience, and The Japan Prize Foundation.
Conflict of interest
The author declares no competing financial interests.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Saito, A. Physiological functions of endoplasmic reticulum stress transducer OASIS in central nervous system. Anat Sci Int 89, 11–20 (2014). https://doi.org/10.1007/s12565-013-0214-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12565-013-0214-x