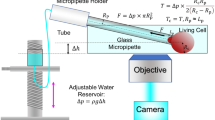
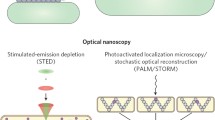
Abstract
The focus of the cell biology field is now shifting from characterizing cellular activities to organelle and molecular behaviors. This process accompanies the development of new biophysical visualization techniques that offer high spatial and temporal resolutions with ultra-sensitivity and low cell toxicity. They allow the biology research community to observe dynamic behaviors from scales of single molecules, organelles, cells to organoids, and even live animal tissues. In this review, we summarize these biophysical techniques into two major classes: the mechanical nanotools like dynamic force spectroscopy (DFS) and the optical nanotools like single-molecule and super-resolution microscopy. We also discuss their applications in elucidating molecular dynamics and functionally mapping of interactions between inter-cellular networks and intra-cellular components, which is key to understanding cellular processes such as adhesion, trafficking, inheritance, and division.



Similar content being viewed by others
References
Alieva, N.O., Efremov, A.K., Hu, S., Oh, D., Chen, Z., Natarajan, M., Ong, H.T., Jegou, A., Romet-Lemonne, G., Groves, J.T., et al. (2017). Force dependence of filopodia adhesion: involvement of myosin II and formins. bioRxiv
Balzarotti F, Eilers Y, Gwosch KC, Gynna AH, Westphal V, Stefani FD, Elf J, Hell SW (2017) Nanometer resolution imaging and tracking of fluorescent molecules with minimal photon fluxes. Science 355:606–612
Bhatia SN, Ingber DE (2014) Microfluidic organs-on-chips. Nat Biotechnol 32:760–772
Butera, D., Passam, F., Ju, L., Cook, K.M., Woon, H., Aponte-Santamaria, C., Gardiner, E., Davis, A.K., Murphy, D.A., Bronowska, A., et al. (2018). Autoregulation of von Willebrand factor function by a disulfide bond switch. Sci Adv 4, eaaq1477
Chaudhuri O, Parekh SH, Lam WA, Fletcher DA (2009) Combined atomic force microscopy and side-view optical imaging for mechanical studies of cells. Nat Meth 6:383–387
Chen W, Lou J, Evans EA, Zhu C (2012) Observing force-regulated conformational changes and ligand dissociation from a single integrin on cells. J Cell Biol 199:497–512
Chen BC, Legant WR, Wang K, Shao L, Milkie DE, Davidson MW, Janetopoulos C, Wu XS, Hammer JA 3rd, Liu Z et al (2014) Lattice light-sheet microscopy: imaging molecules to embryos at high spatiotemporal resolution. Science 346:1257998
Chen Y, Liu B, Ju L, Hong J, Ji Q, Chen W, Zhu C (2015) Fluorescence biomembrane force probe: concurrent quantitation of receptor-ligand kinetics and binding-induced intracellular signaling on a single cell. J Vis Exp. https://doi.org/10.3791/52975
Chen Y, Ju L, Rushdi M, Ge C, Zhu C (2017a) Receptor-mediated cell mechanosensing. Mol Biol Cell 28:3134–3155
Chen Y, Lee H, Tong H, Schwartz M, Zhu C (2017b) Force regulated conformational change of integrin alphaVbeta3. Matrix Biol 60-61:70–85
Chen C, Wang F, Wen S, Su PQ, Wu MCL, Liu Y, Wang B, Li D, Shan X, Kianinia M, Aharonovich I, Toth M, Jackson SP, Xi P, Jin D (2018a) Multi-photon near-infrared emission saturation nanoscopy using upconversion nanoparticles. Nat Commun. https://doi.org/10.1038/s41467-018-05842-w
Chen Y, Su QP, Sun Y, Yu L (2018b) Visualizing autophagic lysosome reformation in cells using in vitro reconstitution systems. Current Protocols Cell Biology 78:11.24.11–11.24.15
Chmyrov A, Keller J, Grotjohann T, Ratz M, d'Este E, Jakobs S, Eggeling C, Hell SW (2013) Nanoscopy with more than 100,000 'doughnuts'. Nat Methods 10:737–740
Das DK, Feng Y, Mallis RJ, Li X, Keskin DB, Hussey RE, Brady SK, Wang JH, Wagner G, Reinherz EL et al (2015) Force-dependent transition in the T-cell receptor beta-subunit allosterically regulates peptide discrimination and pMHC bond lifetime. Proc Natl Acad Sci U S A 112:1517–1522
del Rio A, Perez-Jimenez R, Liu R, Roca-Cusachs P, Fernandez JM, Sheetz MP (2009) Stretching single talin rod molecules activates vinculin binding. Science 323:638–641
Deng W, Xu Y, Chen W, Paul DS, Syed AK, Dragovich MA, Liang X, Zakas P, Berndt MC, Di Paola J et al (2016) Platelet clearance via shear-induced unfolding of a membrane mechanoreceptor. Nat Commun 7:12863
Du W, Su QP, Chen Y, Zhu Y, Jiang D, Rong Y, Zhang S, Zhang Y, Ren H, Zhang C et al (2016) Kinesin 1 drives autolysosome tubulation. Dev Cell 37:326–336
Dulin D, Berghuis BA, Depken M, Dekker NH (2015) Untangling reaction pathways through modern approaches to high-throughput single-molecule force-spectroscopy experiments. Curr Opin Struct Biol 34:116–122
Eisenstein M (2018) Organoids: the body builders. Nat Methods 15:19
Evans E, Leung A, Heinrich V, Zhu C (2004) Mechanical switching and coupling between two dissociation pathways in a P-selectin adhesion bond. Proc Natl Acad Sci 101:11281–11286
Favre-Bulle IA, Stilgoe AB, Rubinsztein-Dunlop H, Scott EK (2017) Optical trapping of otoliths drives vestibular behaviours in larval zebrafish. Nat Commun 8:630
Feng Y, Brazin KN, Kobayashi E, Mallis RJ, Reinherz EL, Lang MJ (2017) Mechanosensing drives acuity of alphabeta T-cell recognition. Proc Natl Acad Sci U S A 114:E8204–E8213
Fiore VF, Ju L, Chen Y, Zhu C, Barker TH (2014) Dynamic catch of a Thy-1–α5β1+syndecan-4 trimolecular complex. Nat Commun 5:4886
Francis EA, Heinrich V (2018) Extension of chemotactic pseudopods by nonadherent human neutrophils does not require or cause calcium bursts. Sci Signal 11(521):eaal4289
Fu H, Jiang Y, Yang D, Scheiflinger F, Wong WP, Springer TA (2017) Flow-induced elongation of von Willebrand factor precedes tension-dependent activation. Nat Commun 8:324
Ghodke H, Caldas VE, Punter CM, van Oijen AM, Robinson A (2016) Single-molecule specific mislocalization of red fluorescent proteins in live Escherichia coli. Biophys J 111:25–27
Guan R, Zhang L, Su QP, Mickolajczyk KJ, Chen GY, Hancock WO, Sun Y, Zhao Y, Chen Z (2017) Crystal structure of Zen4 in the apo state reveals a missing conformation of kinesin. Nat Commun 8:14951
Guo Q, Bishop CJ, Meyer RA, Wilson DR, Olasov L, Schlesinger DE, Mather PT, Spicer JB, Elisseeff JH, Green JJ (2018) Entanglement-based thermoplastic shape memory polymeric particles with photothermal actuation for biomedical applications. ACS Appl Mater Interfaces 10:13333–13341
Hafi N, Grunwald M, van den Heuvel LS, Aspelmeier T, Chen JH, Zagrebelsky M, Schutte OM, Steinem C, Korte M, Munk A et al (2014) Fluorescence nanoscopy by polarization modulation and polarization angle narrowing. Nat Methods 11:579–584
Heller I, Sitters G, Broekmans OD, Farge G, Menges C, Wende W, Hell SW, Peterman EJ, Wuite GJ (2013) STED nanoscopy combined with optical tweezers reveals protein dynamics on densely covered DNA. Nat Methods 10:910–916
Hu KH, Butte MJ (2016) T cell activation requires force generation. J Cell Biol 213:535–542
Huang B, Babcock H, Zhuang X (2010) Breaking the diffraction barrier: super-resolution imaging of cells. Cell 143:1047–1058
Huang X, Fan J, Li L, Liu H, Wu R, Wu Y, Wei L, Mao H, Lal A, Xi P et al (2018) Fast, long-term, super-resolution imaging with Hessian structured illumination microscopy. Nat Biotechnol 36(5):451
Husson J, Chemin K, Bohineust A, Hivroz C, Henry N (2011) Force generation upon T cell receptor engagement. PLoS One 6:e19680
Jia JM, Chowdary PD, Gao X, Ci B, Li W, Mulgaonkar A, Plautz EJ, Hassan G, Kumar A, Stowe AM et al (2017) Control of cerebral ischemia with magnetic nanoparticles. Nat Methods 14:160–166
Jiang N, Huang J, Edwards LJ, Liu B, Zhang Y, Beal CD, Evavold BD, Zhu C (2011) Two-stage cooperative T cell receptor-peptide major histocompatibility complex-CD8 trimolecular interactions amplify antigen discrimination. Immunity 34:13–23
Ju L, Dong J-F, Cruz MA, Zhu C (2013) The N-terminal flanking region of the A1 domain regulates the force-dependent binding of von Willebrand factor to platelet glycoprotein Ibα. J Biol Chem 288:32289–32301
Ju L, Lou J, Chen Y, Li Z, Zhu C (2015) Force-induced unfolding of leucine-rich repeats of glycoprotein Ibα strengthens ligand interaction. Biophys J 109:1781–1784
Ju L, Chen Y, Xue L, Du X, Zhu C (2016) Cooperative unfolding of distinctive mechanoreceptor domains transduces force into signals. ELife 5:e15447
Ju L, Chen Y, Li K, Yuan Z, Liu B, Jackson SP, Zhu C (2017a) Dual biomembrane force probe enables single-cell mechanical analysis of signal crosstalk between multiple molecular species. Sci Rep 7:14185
Ju, L., Chen, Y., Rushdi, M.N., Chen, W., and Zhu, C. (2017b). Two-dimensional analysis of cross-junctional molecular interaction by force probes. In methods Mol Biol: the immune synapse, M.L. Dustin, ed. (springer nature), pp. 231-258
Ju L, McFadyen JD, Al-Daher S, Alwis I, Chen Y, Tonnesen LL, Maiocchi S, Coulter B, Calkin AC, Felner EI et al (2018) Compression force sensing regulates integrin alphaIIbbeta3 adhesive function on diabetic platelets. Nat Commun 9:1087
Kim ST, Takeuchi K, Sun ZY, Touma M, Castro CE, Fahmy A, Lang MJ, Wagner G, Reinherz EL (2009) The alphabeta T cell receptor is an anisotropic mechanosensor. J Biol Chem 284:31028–31037
Kong F, Garcia AJ, Mould AP, Humphries MJ, Zhu C (2009) Demonstration of catch bonds between an integrin and its ligand. J Cell Biol 185:1275–1284
Kong, F., Li, Z., Parks, W.M., Dumbauld, D.W., García, A.J., Mould, A.P., Humphries, M.J., and Zhu, C. (2013). Cyclic mechanical reinforcement of integrin-ligand interactions. Mol Cell
Le Trong I, Aprikian P, Kidd BA, Forero-Shelton M, Tchesnokova V, Rajagopal P, Rodriguez V, Interlandi G, Klevit R, Vogel V et al (2010) Structural basis for mechanical force regulation of the adhesin FimH via finger trap-like β sheet twisting. Cell 141:645–655
Li Z, Kong F, Zhu C (2016) A model for cyclic mechanical reinforcement. Sci Rep 6:35954
Liu B, Chen W, Evavold BD, Zhu C (2014a) Accumulation of dynamic catch bonds between TCR and agonist peptide-MHC triggers T cell signaling. Cell 157:357–368
Liu Z, Xing D, Su QP, Zhu Y, Zhang J, Kong X, Xue B, Wang S, Sun H, Tao Y, Sun Y (2014b) Super-resolution imaging and tracking of protein–protein interactions in sub-diffraction cellular space. Nat Commun 5:4443
Liu B, Chen W, Zhu C (2015a) Molecular force spectroscopy on cells. Annu Rev Phys Chem 66:427–451
Liu Z, Liu Y, Chang Y, Seyf HR, Henry A, Mattheyses AL, Yehl K, Zhang Y, Huang Z, Salaita K (2015b) Nanoscale optomechanical actuators for controlling mechanotransduction in living cells. Nat Meth:1–8
Liu TL, Upadhyayula S, Milkie DE, Singh V, Wang K, Swinburne IA, Mosaliganti KR, Collins ZM, Hiscock TW, Shea J et al (2018) Observing the cell in its native state: imaging subcellular dynamics in multicellular organisms. Science 360(6386):eaaq1392
Lu Y, Zhao J, Zhang R, Liu Y, Liu D, Goldys EM, Yang X, Xi P, Sunna A, Lu J et al (2013) Tunable lifetime multiplexing using luminescent nanocrystals. Nat Photonics 8:32–36
Luca, V.C., Kim, B.C., Ge, C., Kakuda, S., Wu, D., Roein-Peikar, M., Haltiwanger, R.S., Zhu, C., Ha, T., and Garcia, K.C. (2017). Notch-jagged complex structure implicates a catch bond in tuning ligand sensitivity. Science
Luo BH, Springer TA (2006) Integrin structures and conformational signaling. Curr Opin Cell Biol 18:579–586
Matthews BD, Overby DR, Mannix R, Ingber DE (2006) Cellular adaptation to mechanical stress: role of integrins, Rho, cytoskeletal tension and mechanosensitive ion channels. J Cell Sci 119:508–518
Miao Q, Xie C, Zhen X, Lyu Y, Duan H, Liu X, Jokerst JV, Pu K (2017) Molecular afterglow imaging with bright, biodegradable polymer nanoparticles. Nat Biotechnol 35:1102–1110
Neuman KC, Nagy A (2008) Single-molecule force spectroscopy: optical tweezers, magnetic tweezers and atomic force microscopy. Nat Meth 5:491–505
Passam F, Chiu J, Ju L, Pijning A, Jahan Z, Mor-Cohen R, Yeheskel A, Kolsek K, Tharichen L, Aponte-Santamaria C et al (2018) Mechano-redox control of integrin de-adhesion. eLife 7:e34843
Pryshchep S, Zarnitsyna VI, Hong J, Evavold BD, Zhu C (2014) Accumulation of serial forces on TCR and CD8 frequently applied by agonist antigenic peptides embedded in MHC molecules triggers calcium in T cells. J Immunol 193:68–76
Reineck P, Lau DWM, Wilson ER, Nunn N, Shenderova OA, Gibson BC (2018) Visible to near-IR fluorescence from single-digit detonation nanodiamonds: excitation wavelength and pH dependence. Sci Rep 8:2478
Rios AC, Clevers H (2018) Imaging organoids: a bright future ahead. Nat Methods 15:24
Sahl SJ, Hell SW, Jakobs S (2017) Fluorescence nanoscopy in cell biology. Nat Rev Mol Cell Biol 18:685
Sawicka A, Babataheri A, Dogniaux S, Barakat AI, Gonzalez-Rodriguez D, Hivroz C, Husson J (2017) Micropipette force probe to quantify single-cell force generation: application to T-cell activation. Mol Biol Cell 28:3229–3239
Shen M, Zhang N, Zheng S, Zhang WB, Zhang HM, Lu Z, Su QP, Sun Y, Ye K, Li XD (2016) Calmodulin in complex with the first IQ motif of myosin-5a functions as an intact calcium sensor. Proc Natl Acad Sci U S A 113:E5812–e5820
Smith ML, Gourdon D, Little WC, Kubow KE, Eguiluz RA, Luna-Morris S, Vogel V (2007) Force-induced unfolding of fibronectin in the extracellular matrix of living cells. PLoS Biol 5:e268
Somers WS, Tang J, Shaw GD, Camphausen RT (2000) Insights into the molecular basis of leukocyte tethering and rolling revealed by structures of P- and E-selectin bound to SLe(X) and PSGL-1. Cell 103:467–479
Springer TA, Dustin ML (2012) Integrin inside-out signaling and the immunological synapse. Curr Opin Cell Biol 24:107–115
Stephenson NL, Avis JM (2012) Direct observation of proteolytic cleavage at the S2 site upon forced unfolding of the Notch negative regulatory region. Proc Natl Acad Sci U S A 109:E2757–E2765
Su QP, Du W, Ji Q, Xue B, Jiang D, Zhu Y, Lou J, Yu L, Sun Y (2016) Vesicle size regulates nanotube formation in the cell. Sci Rep 6:24002
Sun Y, Schroeder HW 3rd, Beausang JF, Homma K, Ikebe M, Goldman YE (2007) Myosin VI walks “wiggly” on actin with large and variable tilting. Mol Cell 28:954–964
Takebe T, Zhang B, Radisic M (2017) Synergistic engineering: organoids meet organs-on-a-Chip. Cell Stem Cell 21:297–300
Valm AM, Cohen S, Legant WR, Melunis J, Hershberg U, Wait E, Cohen AR, Davidson MW, Betzig E, Lippincott-Schwartz J (2017) Applying systems-level spectral imaging and analysis to reveal the organelle interactome. Nature 546:162–167
Vlachogiannis G, Hedayat S, Vatsiou A, Jamin Y, Fernandez-Mateos J, Khan K, Lampis A, Eason K, Huntingford I, Burke R et al (2018) Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science 359:920–926
Wang C, Du W, Su QP, Zhu M, Feng P, Li Y, Zhou Y, Mi N, Zhu Y, Jiang D et al (2015) Dynamic tubulation of mitochondria drives mitochondrial network formation. Cell Res 25:1108–1120
Wang Y, Botvinick E, Zhao Y, Berns M, Usami S, Tsien R, Chien S (2005) Visualizing the mechanical activation of Src. Nature 434:1040–1045
Wu T, Lin J, Cruz MA, Dong JF, Zhu C (2010) Force-induced cleavage of single VWFA1A2A3 tridomains by ADAMTS-13. Blood 115:370–378
Xu XR, Wang Y, Adili R, Ju L, Spring, CM, Jin, JW, Yang H, Neves MAD, Chen P, Yang Y, Lei X, Chen Y, Gallant RC, Xu M, Zhang H, Song J, Ke P, Zhang D, Carrim N, Yu SY, Zhu G, She YM, Cyr T, Fu W, Liu G, Connelly PW, Rand ML, Adeli K, Freedman J, Lee JE, Tso P, Marchese P, Davidson WS, Jackson SP, Zhu C, Ruggeri ZM, Ni H (2018) Apolipoprotein A-IV binds aIIbß3 integrin and inhibits thrombosis. Nat Commun. https://doi.org/10.1038/s41467-018-05806-0
Yildiz A, Forkey JN, McKinney SA, Ha T, Goldman YE, Selvin PR (2003) Myosin V walks hand-over-hand: single fluorophore imaging with 1.5-nm localization. Science 300:2061–2065
Zhang W, Deng W, Zhou L, Xu Y, Yang W, Liang X, Wang Y, Kulman JD, Zhang XF, Li R (2015) Identification of a juxtamembrane mechanosensitive domain in the platelet mechanosensor glycoprotein Ib-IX complex. Blood 125:562–569
Zhang X, Halvorsen K, Zhang CZ, Wong WP, Springer TA (2009) Mechanoenzymatic cleavage of the ultralarge vascular protein von Willebrand factor. Science 324:1330–1334
Zhanghao K, Chen L, Yang X-S, Wang M-Y, Jing Z-L, Han H-B, Zhang MQ, Jin D, Gao J-T, Xi P (2016) Super-resolution dipole orientation mapping via polarization demodulation. Light: Sci Applications 5:e16166–e16166
Zhao J, Jin D, Schartner EP, Lu Y, Liu Y, Zvyagin AV, Zhang L, Dawes JM, Xi P, Piper JA et al (2013) Single-nanocrystal sensitivity achieved by enhanced upconversion luminescence. Nat Nanotechnol 8:729–734
Zhong MC, Gong L, Zhou JH, Wang ZQ, Li YM (2013a) Optical trapping of red blood cells in living animals with a water immersion objective. Opt Lett 38:5134–5137
Zhong MC, Wei XB, Zhou JH, Wang ZQ, Li YM (2013b) Trapping red blood cells in living animals using optical tweezers. Nat Commun 4:1768
Zong W, Wu R, Li M, Hu Y, Li Y, Li J, Rong H, Wu H, Xu Y, Lu Y et al (2017) Fast high-resolution miniature two-photon microscopy for brain imaging in freely behaving mice. Nat Methods 14:713–719
Zong W, Zhao J, Chen X, Lin Y, Ren H, Zhang Y, Fan M, Zhou Z, Cheng H, Sun Y et al (2015) Large-field high-resolution two-photon digital scanned light-sheet microscopy. Cell Res 25:254–257
Acknowledgments
We thank Shaun P. Jackson, Dayong Jin, and their labs for helpful discussion.
Author information
Authors and Affiliations
Contributions
Q.P.S. and L.A.J. contributed equally, prepared figures, and wrote and edited the manuscript together.
Corresponding authors
Ethics declarations
Funding
This work was supported by National Heart Foundation of Australia Postdoctoral Fellowship 101285 (LJ), Sydney Local Health District - Annual Health Research Infrastructure Award (LJ), The Royal College of Pathologists of Australasia Kanematsu Research Award (LJ).
Conflict of interest
Qian Peter Su declares that he has no conflict of interest. Lining Arnold Ju declares that he has no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Rights and permissions
About this article
Cite this article
Su, Q.P., Ju, L.A. Biophysical nanotools for single-molecule dynamics. Biophys Rev 10, 1349–1357 (2018). https://doi.org/10.1007/s12551-018-0447-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12551-018-0447-y