Abstract
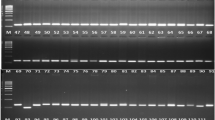
Genetic diversity was assessed among 53 Indian garlic accessions using SSR markers. Initially, 24 SSR primer pairs were used for screening three selected garlic accessions. Out of 24 SSR primer pairs, 10 primer pairs which consistently showed good amplification and polymorphism were selected for DNA profiling. SSR primer pairs showed PIC values ranging from 0.30 to 0.99. Based on AMOVA we found that the greater part of the genetic diversity was expected due to intra population with 84% variation and only 16% of variation was due to among populations suggesting presence of genetic structure. The results of cluster analysis and principal component analysis largely correspond to each other. Population structure analysis revealed genetic differentiation of accessions. The results of present study revealed existence of significant variability in Indian garlic germplasm.



Similar content being viewed by others
References
Abdoli M, Habibi-Khaniani B, Baghalian K, Shahnazi S, Rassouli H (2009) Classification of Iranian garlic (A. sativum L.) ecotypes using RAPD marker. J Med Plants 8:45–51
Avato P, Miccolis V, Tursi F (1998) Agronomic evaluation and essential oil content of garlic (Allium sativum L.) ecotypes grown in southern Italy. Adv Hort Sci 4:201–204
Baghalian K, Ziai SA, Naghavi MR, Badi HN, Khalighi A (2005) Evaluation of allicin content and botanical traits in Iranian garlic (Allium sativum L.) ecotypes. Sci Hortic 103:155–166
Barboza K, Beretta V, Kozub PC et al (2018) Microsatellite analysis and marker development in garlic: distributionin EST sequence, genetic diversity analysis, and marker transferability across Alliaceae. Mol Gen Genom. https://doi.org/10.1007/s00438-018-1442-5
Botstein D, White RL, Skolnick M, Davis RW (1980) Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet 32:314–331
Bradley KF, Rieger MA, Collins GG (1996) Classification of Australian garlic cultivars by DNA fingerprinting. Aust J Exp Agric 36:613–618
Chen S, Zhou J, Chen Q, Chang Y, Du J, Meng H (2013) Analysis of the genetic diversity of garlic (Allium sativum L.) germplasm by SRAP. Biochem Syst Ecol 50:139–146
Chen S, Chen W, Shen X, Yang Y, Qi F, Liu Y (2014) Analysis of the genetic diversity of garlic (Allium sativum L.) by simple sequence repeat and inter simple sequence repeat analysis and agro-morphological traits. Biochem Syst Ecol 55:260–267
Cunha CP, Hoogerheide ES, Zucchi MI, Monteiro M, Pinheiro JB (2012) New microsatellite markers for garlic, Allium sativum (Alliaceae). Am J Bot 99:e17–e19
Cunha CP, Resende FV, Zucchi MI, Pinheiro JB (2014) SSR-based genetic diversity and structure of garlic accessions from Brazil. Genetica 142:419–431
Doyal JJ, Doyal JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Earl DA, von Holdt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Res 4:359–361
Etoh T, Simon PW (2002) Diversity, fertility and seed production of garlic. In: Rabinowitch HD, Currah L (eds) Allium crop science: recent advances. CAB International, New York, pp 101–117
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Excoffier L (1993) Analysis of molecular variance (AMOVA) version 1.55. In: Genetics and Biometry Laboratory, University of Geneva, Switzerland
Excoffier L, Smouse PE, Quattro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction sites. Genetics 131:479–491
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587
Figliuolo G, Candido V, Logozzo G, Miccolis V, Spagnoletti PL, Zeuli PL (2001) Genetic evaluation of cultivated garlic germplasm (Allium sativum L. and A. ampeloprasum L.). Euphytica 121:325–334
Gous M, Rafii MY, Ismail MR, Puteh AB, Rahim HA, Islam KN, Latif MA (2013) A review of microsatellite markers and their applications in rice breeding programs to improve blast disease resistance. Int J Mol Sci 14:22499–22528
Gupta VP, Rathore PK, Plaha P (1992) Divergence analysis of some powdery mildew resistance lines of pea. Crop Improv 19:18–22
Gupta PK, Baliyan HS, Sharma PC, Ramesh B (1996) Microsatellite in plants: a new class molecular markers. Curr Sci 70:45–54
Ipek M, Ipek A, Simon PW (2003) Comparison of AFLPs, RAPD markers, and isozymes for diversity assessment of garlic and detection of putative duplicates in germplasm collection. J Am Soc Hortic Sci 128:46–252
Ipek M, Ipek AS, Almquist G, Simon PW (2005) Demonstration of linkage and development of the first low-density genetic map of garlic, based on AFLP markers. Theor Appl Genet 110:228–236
Ipek M, Ipek A, Simon PW (2008) Rapid characterization of garlic clones with locus specific DNA markers. Turk J Agric Sci 32:357–362
Jabbes N, Geoffriau E, Le Clerc V, Dridi B, Hannechi C (2011) Inter simple sequence repeat fingerprints for assess genetic diversity of Tunisian garlic populations. J Agric Sci 3:77–85
Jaccard P (1908) Nouvelles recherché sur la distribution florale. Bull Soc Vaud Sci Nat 44:223–270
Jenderek MM, Hannan RM (2004) Variation in reproductive characteristics and seed production in the USDA garlic germplasm collections. HortScience 39:48–488
Jo MH, Ham I, Moe KT, Kwon SW, Lu FH, Park YJ, Kim WS, Won MK, Kim T, Lee EM (2012) Classification of genetic variation in garlic (Allium sativum L.) using SSR markers. Aust J Crop Sci 6:25–631
Kamenetsky R (2007) Garlic: botany and horticulture. Hortic Rev 33:123–172
Khar A, Devi AA, Lawande KE (2008) Analysis of genetic diversity among Indian garlic (Allium sativum L.) cultivars and breeding lines using RAPD markers. Indian J Genet Plant Breed 68:52–57
Kumar V, Sharma S, Sharma AK, Sharma S, Bhat KV (2009) Comparative analysis of diversity based on morpho-agronomic traits and microsatellite markers in common bean. Euphytica 170:249–262
Lampasona GS, Martınez L, Burba JL (2003) Genetic diversity among selected Argentinean garlic clones (Allium sativum L.) using AFLP (Amplified Fragment Length Polymorphism) Source. Euphytica 132:115–119
Lampasona GS, Asprelli P, Burba JL (2012) Genetic analysis of a garlic (Allium sativum L.) germplasm collection from Argentina. Sci Hortic 138:183–189
Ma KH, Kwag JG, Zhao W, Dixit A, Lee GA, Kim HH, Chung IM, Kim NS, Lee JS, Ji JJ, Kim TS, Park YJ (2009) Isolation and characteristics of eight novel polymorphic microsatellite loci from the genome of garlic (Allium sativum L.). Sci Hortic 122:355–361
Milbourne D, Meyer R, Bradshaw JE, Baird E, Bonar N, Provan J, Powell W, Waugh R (1997) Comparison of PCR-based marker systems for the analysis of genetic relationships in cultivated potato. Mol Breed 3:127–136
Panthee DR, Kc RB, Regmi HN, Subedi PP, Bhattarai S, Dhakal J (2006) Diversity analysis of garlic (Allium sativum L.) germplasms available in Nepal based on morphological characters. Genet Resour Crop Evol 53:205–212
Paredes CM, Becerra VV, Gonzalez AMI (2008) Low genetic diversity among garlic (Allium sativum L.) accessions detected using random amplified polymorphic DNA (RAPD). Chil J Agric Res 68:3–12
Park YJ, Lee JK, Kim NS (2009) Simple sequence repeat polymorphisms (SSRPs) for evaluation of molecular diversity and germplasm classification of minor crops. Molecules 14:4546–4569
Peakall R, Smouse PE (2006) GENALEX6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Pooler MR, Simon PW (1993) Characterization and classification of isozyme and morphological variation in a diverse collection of garlic clones. Euphytica 68:121–130
Prevost A, Wilkinson M (1999) A new system of comparing PCR primers applied to ISSR fingerprinting of potato cultivars. Theor Appl Genet 98:107–112
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Rafalski JA, Vogel JM, Morgante M, Powel W, Andre C, Tingey SV (1996) Generating and using DNA markers in plants. In: Birren B, Lai E (eds) Non mammalian genomics analysis. A practical guide. Academic Press, San Diego, pp 75–134
Ranjekar PK, Pallotta D, Lafontaine JG (1978) Analysis of plant genomes V. Comparative study of molecular properties of DNAs of seven Allium species. Biochem Genet 16:957–970
Rohlf FJ (1998) NTSYSpc. Numerical taxonomy and multivariate analysis system. Version 2.20e, Exeter Software, New York
Shaaf S, Sharma R, Kilian B, Walthe A, Ozkan H, Karami E, Mohammadi B (2014) Genetic structure and eco-geographical adaptation of garlic landraces (Allium sativum L.) in Iran. Genet Res Crop Evol 61:1565–1580
Singh RK, Dubey BK, Gupta RP (2012) Studies on variability and genetic divergence in elite lines of garlic (Allim sativum L.). J Spices Arom Crops 21:136–144
Taucher J, Hansel A, Jordan A, Lindinger W (1996) Analysis of compounds in human breath after ingestion of garlic using proton-transfer-reaction mass spectrometry. J Agric Food Chem 44:3778–3782
Tucak M, Cupic T, Popovic S, Stjepanovic M, Gantner R, Meglic V (2009) Agronomic evaluation and utilization of red clover (Trifolium pratense L.) germplasm. Not Bot Horti Agrobot Cluj Napoca 37:206–210
Volk GM, Henk AD, Richards CM (2004) Genetic diversity among U.S. garlic clones as detected using AFLP methods. J Am Soc Hortic Sci 129:559–569
Wang H, Li X, Shen D, Oiu Y, Song J (2014) Diversity evaluation of morphological traits and allicin content in garlic (Allium sativum L.) from China. Euphytica 198:243–254
Zewdie Y, Havey MJ, Prince JP, Jenderek MM (2005) The first genetic linkages among expressed regions of the garlic genome. J Am Soc Hortic Sci 130(4):569–574
Zhao WG, Chung JW, Lee GA, Ma KH, Kim HH, Kim KT, Chung IM, Lee JK, Kim NS, Kim SM, Park YJ (2011) Molecular genetic diversity and population structure of a selected core set in garlic and its relatives using novel SSR markers. Plant Breed 130:46–54
Zohary D, Hopf M (2012) Domestication of plants in the Old World, 4th edn. Oxford University Press, New York
Acknowledgements
The authors are grateful to Prof. Gaya Prasad, Hon’ble Vice Chancellor of the Sardar Vallabhbhai Patel University of Agriculture and Technology, Meerut, U.P. India for providing facilities and encouragement. Authors are also thankful to PAU; Ludhiana, NHRDF, Karnal and DOGR, Maharashtra for providing garlic material.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Kumar, M., Rakesh Sharma, V., Kumar, V. et al. Genetic diversity and population structure analysis of Indian garlic (Allium sativum L.) collection using SSR markers. Physiol Mol Biol Plants 25, 377–386 (2019). https://doi.org/10.1007/s12298-018-0628-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12298-018-0628-y