Abstract
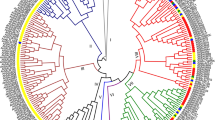
The early auxin responsive SAUR family is an important gene family in auxin signal transduction. We here present the first report of a genome-wide identification of SAUR genes in watermelon genome. We successfully identified 65 ClaSAURs and provide a genomic framework for future study on these genes. Phylogenetic result revealed a Cucurbitaceae-specific SAUR subfamily and contribute to understanding of the evolutionary pattern of SAUR genes in plants. Quantitative RT-PCR analysis demonstrates the existed expression of 11 randomly selected SAUR genes in watermelon tissues. ClaSAUR36 was highly expressed in fruit, for which further study might bring a new prospective for watermelon fruit development. Moreover, correlation analysis revealed the similar expression profiles of SAUR genes between watermelon and Arabidopsis during shoot organogenesis. This work gives us a new support for the conserved auxin machinery in plants.





Similar content being viewed by others
References
Altschul SF, Madden TL, Schaffer AA, Zhang J et al (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucl Acids Res 25:3389–3402
Bailey TL, Boden M, Buske FA, Frith M et al (2009) MEME SUITE: tools for motif discovery and searching. Nucl Acids Res 37:W202–W208
Bao Y, Dharmawardhana P, Mockler TC, Strauss SH (2009) Genome scale transcriptome analysis of shoot organogenesis in Populus. BMC Plant Biol 9:132
Chae K, Isaacs CG, Reeves PH, Maloney GS et al (2012) Arabidopsis SMALL AUXIN UP RNA63 promotes hypocotyl and stamen filament elongation. Plant J 71:684–697
Che P, Lall S, Nettleton D, Howell SH (2006) Gene expression programs during shoot, root, and callus development in Arabidopsis tissue culture. Plant Physiol 141:620–637
Chen Y, Hao X, Cao J (2014) Small auxin upregulated RNA (SAUR) gene family in maize: identification, evolution, and its phylogenetic comparison with Arabidopsis, rice, and sorghum. J Integr Plant Biol 56:133–150
Choi PS, Soh WY, Kim YS, Yoo OJ et al (1994) Genetic transformation and plant regeneration of watermelon using Agrobacterium tumefaciens. Plant Cell Rep 13:344–348
Collins JK, Wu GY, Perkins-Veazie P, Spears K et al (2007) Watermelon consumption increases plasma arginine concentrations in adults. Nutrition 23:261–266
De Smet I, Voss U, Lau S, Wilson M et al (2011) Unraveling the evolution of auxin signaling. Plant Physiol 155:209–221
Duclercq J, Sangwan-Norreel B, Catterou M, Sangwan RS (2011) De novo shoot organogenesis: from art to science. Trends Plant Sci 16:597–606
Finet C, Jaillais Y (2012) AUXOLOGY: when auxin meets plant evo-devo. Dev Biol 369:19–31
Gil P, Liu Y, Orbovic V, Verkamp E et al (1994) Characterization of the auxin-inducible SAUR-AC1 gene for use as a molecular genetic tool in Arabidopsis. Plant Physiol 104:777–784
Guo SG, Liu JA, Zheng Y, Huang MY et al (2011) Characterization of transcriptome dynamics during watermelon fruit development: sequencing, assembly, annotation and gene expression profiles. BMC Genom 12:454
Guo S, Zhang J, Sun H, Salse J et al (2013) The draft genome of watermelon (Citrullus lanatus) and resequencing of 20 diverse accessions. Nat Genet 45:51–58
Hanada K, Zou C, Lehti-Shiu MD, Shinozaki K et al (2008) Importance of lineage-specific expansion of plant tandem duplicates in the adaptive response to environmental stimuli. Plant Physiol 148:993–1003
Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucl Acids Res 27:297–300
Huang S, Li R, Zhang Z, Li L et al (2009) The genome of the cucumber, Cucumis sativus L. Nat Genet 41:1275–1281
Huang X, Chen J, Bao Y, Liu L et al (2014) Transcript profiling reveals auxin and cytokinin signaling pathways and transcription regulation during in vitro organogenesis of ramie (Boehmeria nivea L. Gaud). PLoS ONE 9:e113768
Huang X, Bao Y, Wang B, Liu L et al (2016) Identification of small auxin-up RNA (SAUR) genes in Urticales plants: mulberry (Morus notabilis), hemp (Cannabis sativa) and ramie (Boehmeria nivea). J Genet 95(1):119–129
Jain M, Tyagi AK, Khurana JP (2006) Genome-wide analysis, evolutionary expansion, and expression of early auxin-responsive SAUR gene family in rice (Oryza sativa). Genomics 88:360–371
Kong Q, Yuan J, Gao L, Zhao S et al (2014) Identification of suitable reference genes for gene expression normalization in qRT-PCR analysis in watermelon. PLoS ONE 9:e90612
Lespinet O, Wolf YI, Koonin EV, Aravind L (2002) The role of lineage-specific gene family expansion in the evolution of eukaryotes. Genome Res 12:1048–1059
Liu RX, Kuang J, Gong Q, Hou XL (2003) Principal component regression analysis with SPSS. Comput Meth Prog Biomed 71:141–147
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2 (-Delta Delta C(T)) Method. Methods 25:402–408
Matsui K, Hiratsu K, Koyama T, Tanaka H et al (2005) A chimeric AtMYB23 repressor induces hairy roots, elongation of leaves and stems, and inhibition of the deposition of mucilage on seed coats in Arabidopsis. Plant Cell Physiol 46:147–155
McClure BA, Hagen G, Brown CS, Gee MA et al (1989) Transcription, organization, and sequence of an auxin-regulated gene cluster in soybean. Plant Cell 1:229–239
Ouibrahim L, Mazier M, Estevan J, Pagny G et al (2014) Cloning of the Arabidopsis rwm1 gene for resistance to Watermelon mosaic virus points to a new function for natural virus resistance genes. Plant J 79:705–716
Paponov IA, Paponov M, Teale W, Menges M et al (2008) Comprehensive transcriptome analysis of auxin responses in Arabidopsis. Mol Plant 1:321–337
Pattison RJ, Csukasi F, Catalá C (2014) Mechanisms regulating auxin action during fruit development. Physiol Plant 151(1):62–72
Roig-Villanova I, Bou-Torrent J, Galstyan A, Carretero-Paulet L et al (2007) Interaction of shade avoidance and auxin responses: a role for two novel atypical bHLH proteins. EMBO J 26:4756–4767
Roux C, Bilang J, Theunissen BH, Perrot-Rechenmann C (1998) Identification of new early auxin markers in tobacco by mRNA differential display. Plant Mol Biol 37:385–389
Sato A, Sasaki S, Matsuzaki J, Yamamoto KT (2014) Light-dependent gravitropism and negative phototropism of inflorescence stems in a dominant Aux/IAA mutant of Arabidopsis thaliana, axr2. J Plant Res 127:627–639
Spartz AK, Lee SH, Wenger JP, Gonzalez N et al (2012) The SAUR19 subfamily of SMALL AUXIN UP RNA genes promote cell expansion. Plant J 70:978–990
Spartz AK, Ren H, Park MY, Grandt KN et al (2014) SAUR Inhibition of PP2C-D Phosphatases Activates Plasma Membrane H + -ATPases to Promote Cell Expansion in Arabidopsis. Plant Cell 26:2129–2142
Stamm P, Kumar PP (2013) Auxin and gibberellin responsive Arabidopsis SMALL AUXIN UP RNA36 regulates hypocotyl elongation in the light. Plant Cell Rep 32:759–769
Tamura K, Peterson D, Peterson N, Stecher G et al (2011) MEGA5: molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol Biol Evol 28:2731–2739
Wu J, Liu S, He Y, Guan X et al (2012) Genome-wide analysis of SAUR gene family in Solanaceae species. Gene 509:38–50
Yu CS, Chen YC, Lu CH, Hwang JK (2006) Prediction of protein subcellular localization. Proteins 64:643–651
Zhang N, Huang X, Bao Y, Wang B et al (2015) Genome-wide identification and expression profiling of WUSCHEL-related homeobox (WOX) genes during adventitious shoot regeneration of watermelon (Citrullus lanatus). Acta Physiol Plant 37:224
Acknowledgements
We would like to thank Dr. Adel M. R. A. Abdelaziz from Central Laboratory of Organic Agriculture, Agricultural Research Center (Giza 12619, Egypt) for manuscript revision. This study was supported by Applied Basic Research Project of Wuhan City (2015021701011611), Talent Project for Wuhan Institute of Agricultural Science (CX201615-06) and Supporting Program for Science and Technology Research of Hubei Province (2015BBA201).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhang, N., Huang, X., Bao, Y. et al. Genome-wide identification of SAUR genes in watermelon (Citrullus lanatus). Physiol Mol Biol Plants 23, 619–628 (2017). https://doi.org/10.1007/s12298-017-0442-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12298-017-0442-y