Abstract
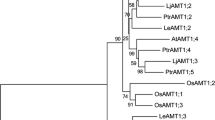
Uroporphyrinogen III methyl transferase (UPM1) and Sirohydrochlorin ferrochelatase (SIRB) are the important genes involved in the biosynthesis of siroheme, the prosthetic group of nitrite reductases (NiR) and sulfite reductases (SiR) involved in nitrogen and sulfur assimilation. Both UPM1 and SIRB could be potential candidate genes targeted for sustainable agriculture especially in N-deficient soil. The phylogenetic analysis revealed that these genes are highly conserved among algae, bryophytes and vascular plants including dicots and monocots. The Arabidopsis proteins UPM1 and SIRB have close similarity with Camelina sativa followed by Brassica napus, Brassica rapa, and Brassica oleracea of the family brassicaceae. The tissue specific expression studies revealed that both the gene are expressed in stem, flower and silique and have highest expression in leaves where the protein content is quite high. The in silico promoter analysis revealed the presence of several light-responsive elements like GATA box, G box, I box, SORLIP2, SORLIP5, SORLREP3 and SORLREP4. Therefore, expression of both the genes was minimal in etiolated seedlings and was upregulated in light. Photo-regulation of transcript abundance of UPM1 and SIRB involved in the biosynthesis of siroheme the cofactor involved in 6 electron reduction of NO2 − and SO 2−3 by NiR and SiR is crucial as the gene expression of latter two enzymes along with other N and S assimilatory enzymes are also modulated by light.




Similar content being viewed by others
References
Becker TW, Foyer C, Caboche M (1992) Light-regulated expression of the nitrate-reductase and nitrite-reductase genes in tomato and in the phytochrome-deficient urea mutant of tomato. Planta 188:39–47
Behringer C, Schwechheimer C (2015) B-GATA transcription factors—insights into their structure, regulation, and role in plant development Frontiers in. Plant Sci 6:90
Bi YM, Wang RL, Zhu T, Rothstein SJ (2007) Global transcription profiling reveals differential responses to chronic nitrogen stress and putative nitrogen regulatory components in Arabidopsis. BMC Genom 8:281
Brunold C (1993) Regulatory interactions between sulfate and nitrate assimilation. Sulfur nutrition and sulfur assimilation in higher plants. SPB Academic Publishing, The Hague
Cashmore AEMaAR (1994) Isolation and characterization of a fourth Arabidopsis thaliana G-box-binding factor, which has similarities to Fos oncoprotein. Proc Natl Acad Sci USA 91:i–vii
de Cires A, de la Torre A, Delgado B, Lara C (1993) Role of light and CO2 fixation in the control of nitrate-reductase activity in barley leaves. Planta 190:277–283
Deng M-D, Moureaux T, Leydecker MT, Caboche M (1990) Nitrate-reductase expression is under the control of a circadian rhythm and is light inducible in Nicotiana tabacum leaves. Planta 180:257–261
Dutta S, Mohanty S, Tripathy BC (2009) Role of temperature stress on chloroplast biogenesis and protein import in pea. Plant Physiol 150:1050–1061
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39(4):783–791
Giuliano G, Pichersky E, Malik VS, Timko MP, Scolnik PA, Cashmore AR (1988) An evolutionarily conserved protein binding sequence upstream of a plant light-regulated gene. Proc Natl Acad Sci USA 85:7089–7093
Grob U, Stuber K (1987) Discrimination of phytochrome dependent light inducible from non-light inducible plant genes. Prediction of a common light-responsive element (LRE) in phytochrome dependent light inducible plant genes. Nucleic Acids Res 15:9957–9973
Guohua X, Xiaorong F, Anthony JM (2012) Plant nitrogen assimilation and use efficiency. Annu Rev Plant Biol 63:153–182. doi:10.1146/annurev-arplant-042811-105532
Hell RSJ, Bork C (1997) Light and sulfur sources modulate mRNA levels of several genes of sulfate assimilation. Sulfur metabolism in higher plants. Backhuys Publishers, Leiden
Huber SC, Huber JL, Campbell WH, Redinbaugh MG (1992) Comparative studies of the light modulation of nitrate reductase and sucrose-phosphate synthase activities in spinach leaves. Plant Physiol 100:706–712
Hudson ME, Quail PH (2003) Identification of promoter motifs involved in the network of phytochrome a-regulated gene expression by combined analysis of genomic sequence and microarray data. Plant Physiol 133:1605–1616
Kocsy G, Owttrim G, Brander K, Brunold C (1997) Effect of chilling on the diurnal rhythm of enzymes involved in protection against oxidative stress in a chilling-tolerant and a chilling-sensitive maize genotype. Physiol Plant 99:249–254
Kopriva S, Muheim R, Koprivova A, Trachsel N, Catalano C, Suter M, Brunold C (1999) Light regulation of assimilatory sulphate reduction in Arabidopsis thaliana. Plant J 20:37–44
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆Ct method. Methods 25:402–408
Menkens AE, Schindler U, Cashmore AR (1995) The G-box: a ubiquitous regulatory DNA element in plants bound by the GBF family of bZIP proteins. Trends Biochem Sci 20:506–510
Murphy MJ, Siegel LM, Tove SR, Kamin H (1974) Siroheme: a new prosthetic group participating in six-electron reduction reactions catalyzed by both sulfite and nitrite reductases. Proc Natl Acad Sci USA 71:612–616
Parry MAJ, Andralojc PJ, Mitchell RAC, Madgwick PJ, Keys AJ (2003) Manipulation of Rubisco: the amount, activity, function and regulation. J Exp Bot 54:1321–1333
Puente P, Wei N, Deng XW (1996) Combinatorial interplay of promoter elements constitutes the minimal determinants for light and developmental control of gene expression in Arabidopsis. EMBO J 15:3732–3743
Raghuram N, Chandok MR, Sopory SK (1999) Light regulation of nitrate reductase gene expression in maize involves a G-protein. Mol Cell Biol Res Commun 2:86–90
Rajasekhar VK, Mohr H (1986) Appearance of nitrite reductase in cotyledons of the mustard (Sinapis alba L.) seedling as affected by nitrate, phytochrome and photooxidative damage of plastids. Planta 168:369–376
Rajasekhar VK, Gowri G, Campbell WH (1988) Phytochrome-mediated light regulation of nitrate reductase expression in squash cotyledons. Plant Physiol 88:242–244
Robertson GP, Vitousek PM (2009) Nitrogen in agriculture: balancing the cost of an essential resource. Annu rev environ resour 34:97–125
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Tripathy BC, Sherameti I, Oelmuller R (2010) Siroheme: an essential component for life on earth. Plant Signal Behav 5:14–20
William BT, Anthony RC (1995) Light-regulated transcription. Ann Rev Plant Physiol Plant Mol Biol 46:445–474
Zuckerkandl EPL (1965) Evolutionary divergence and convergence in proteins. Evolving genes and proteins. Academic Press, New York
Acknowledgments
The work was supported by J C Bose fellowship to BCT from the Department of Science and Technology, Government of India.
Authors’ contribution
BCT designed the experiments. SG and NCJ performed the experiments and analyzed the data. SG and BCT prepared the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Sampurna Garai and Naveen Chandra Joshi have contributed equally to this work.
Rights and permissions
About this article
Cite this article
Garai, S., Joshi, N.C. & Tripathy, B.C. Phylogenetic analysis and photoregulation of siroheme biosynthesis genes: uroporphyrinogen III methyltransferase and sirohydrochlorin ferrochelatase of Arabidopsis thaliana . Physiol Mol Biol Plants 22, 351–359 (2016). https://doi.org/10.1007/s12298-016-0363-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12298-016-0363-1