Abstract
Background
Mitochondrial dysfunction can be associated with genomic instability and has been implicated in the pathogenesis of several human malignancies, including breast cancer (BC). The mitochondrial protein, translocase of inner mitochondrial membrane 17 homolog A (TIMM17A) contributes to a pre-protein import complex, essential for mitochondrial function. In this study, TIMM17A mRNA expression was evaluated in benign and malignant breast tissues and correlated with pathological and clinical outcomes.
Methods
Breast cancer tissues (n = 127) and normal tissues (n = 33) underwent RNA extraction and reverse transcription, transcript levels were determined by using real-time quantitative PCR and normalized against CK19. Transcript levels were compared and then analysed against tumour size, tumour grade, oestrogen receptor (ER) status, nodal involvement, TNM stage, Nottingham prognostic index (NPI) and clinical outcome over a 10-year follow-up period.
Results
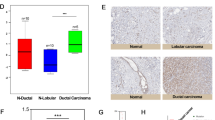
Compared to normal tissue (mean transcript level = 12.7), TIMM17A mRNA expression was higher in BC (62, p = 0.006), TNM-1 (37.9, p = 0.05), TNM-2 (114, p = 0.034), NPI-2 (81, p = 0.041), patients with progressive disease (169, p = 0.017) and those who died from BC (179, p = 0.026). Expression increased with tumour grade; grade 1 versus 2 (12.1 vs.70, p = 0.007), grade 1 versus 3 (12.1 vs.73, p = 0.065, [not significant] NS) and grade 1 versus 2 and 3 (12.1 vs. 72, p = 0.0048). Higher transcript levels were associated with ER-α positivity (94 vs. 31.6, p = 0.073, NS) and ER-β negativity (82 vs. 16, p = 0.015). Nodal positivity was significantly associated with higher transcript levels (101 vs. 28.1, p = 0.046). Compared to disease-free patients (mean transcript level = 24.6), TIMM17A expression was significantly higher in those with progressive disease and patients who died of BC (179, p = 0.037). Higher transcript levels were significantly associated with poorer overall survival after a median follow-up of 10 years (p = 0.010). TIMM17A expression emerged as a strong independent predictor of overall survival in multivariate analysis (p = 0.033).
Conclusion
TIMM17A mRNA expression is significantly associated with unfavourable pathological parameters including tumour grade, nodal positivity, TNM stage and NPI; in addition to adverse clinical outcomes such as progressive disease, disease-free and overall survival. TIMM17A offers utility as a prognostic marker and a novel mitochondrial target for potential therapeutic strategies.

Similar content being viewed by others
References
Birch-Machin MA. The role of mitochondria in ageing and carcinogenesis. Clin Exp Dermatol. 2006;31:548–52.
Rassow J, Dekker PJ, van Wilpe S, Meijer M, Soll J. The preprotein translocase of the mitochondrial inner membrane: function and evolution. J Mol Biol. 1999;286:105–20.
Jensen RE, Dunn CD. Protein import into and across the mitochondrial inner membrane: role of the TIM23 and TIM22 translocons. Biochim Biophys Acta. 2002;1592:25–34.
Neupert W, Herrmann JM. Translocation of proteins into mitochondria. Annu Rev Biochem. 2007;76:723–49.
Rehling P, Brandner K, Pfanner N. Mitochondrial import and the twin-pore translocase. Nat Rev Mol Cell Biol. 2004;5:519–30.
Mokranjac D, Sichting M, Popov-Celeketic D, Mapa K, Gevorkyan-Airapetov L, Zohary K, et al. Role of Tim50 in the transfer of precursor proteins from the outer to the inner membrane of mitochondria. Mol Biol Cell. 2009;20:1400–7.
Popov-Celeketic D, Mapa K, Neupert W, Mokranjac D. Active remodelling of the TIM23 complex during translocation of preproteins into mitochondria. EMBO J. 2008;27:1469–80.
Geissler A, Chacinska A, Truscott KN, Wiedemann N, Brandner K, Sickmann A, et al. The mitochondrial presequence translocase: an essential role of Tim50 in directing preproteins to the import channel. Cell. 2002;111:507–18.
Mokranjac D, Paschen SA, Kozany C, Prokisch H, Hoppins SC, Nargang FE, et al. Tim50, a novel component of the TIM23 preprotein translocase of mitochondria. EMBO J. 2003;22:816–25.
Neupert W, Brunner M. The protein import motor of mitochondria. Nat Rev Mol Cell Biol. 2002;3:555–65.
Moro F, Okamoto K, Donzeau M, Neupert W, Brunner M. Mitochondrial protein import: molecular basis of the ATP-dependent interaction of MtHsp70 with Tim44. J Biol Chem. 2002;277:6874–80.
Mokranjac D, Popov-Celeketic D, Hell K, Neupert W. Role of Tim21 in mitochondrial translocation contact sites. J Biol Chem. 2005;280:23437–40.
Albrecht R, Rehling P, Chacinska A, Brix J, Cadamuro SA, Volkmer R, et al. The Tim21 binding domain connects the preprotein translocases of both mitochondrial membranes. EMBO Rep. 2006;7:1233–8.
Chacinska A, van der LM, Mehnert CS, Guiard B, Mick DU, Hutu DP, et al. Distinct forms of mitochondrial TOM-TIM supercomplexes define signal-dependent states of preprotein sorting. Mol Cell Biol. 2010;30:307–18.
Schiller D. Pam17 and Tim44 act sequentially in protein import into the mitochondrial matrix. Int J Biochem Cell Biol. 2009;41:2343–9.
Hutu DP, Guiard B, Chacinska A, Becker D, Pfanner N, Rehling P, et al. Mitochondrial protein import motor: differential role of Tim44 in the recruitment of Pam17 and J-complex to the presequence translocase. Mol Biol Cell. 2008;19:2642–9.
Hoogenraad NJ, Ward LA, Ryan MT. Import and assembly of proteins into mitochondria of mammalian cells. Biochim Biophys Acta. 2002;1592:97–105.
Ryan KR, Menold MM, Garrett S, Jensen RE. SMS1, a high-copy suppressor of the yeast mas6 mutant, encodes an essential inner membrane protein required for mitochondrial protein import. Mol Biol Cell. 1994;5:529–38.
Milenkovic D, Muller J, Stojanovski D, Pfanner N, Chacinska A. Diverse mechanisms and machineries for import of mitochondrial proteins. Biol Chem. 2007;388:891–7.
Bomer U, Meijer M, Maarse AC, Honlinger A, Dekker PJ, Pfanner N, et al. Multiple interactions of components mediating preprotein translocation across the inner mitochondrial membrane. EMBO J. 1997;16:2205–16.
Milisav I, Moro F, Neupert W, Brunner M. Modular structure of the TIM23 preprotein translocase of mitochondria. J Biol Chem. 2001;276:25856–61.
Bauer MF, Gempel K, Reichert AS, Rappold GA, Lichtner P, Gerbitz KD, et al. Genetic and structural characterization of the human mitochondrial inner membrane translocase. J Mol Biol. 1999;289:69–82.
Martinez-Caballero S, Grigoriev SM, Herrmann JM, Campo ML, Kinnally KW. Tim17p regulates the twin pore structure and voltage gating of the mitochondrial protein import complex TIM23. J Biol Chem. 2007;282:3584–93.
Meier S, Neupert W, Herrmann JM. Conserved N-terminal negative charges in the Tim17 subunit of the TIM23 translocase play a critical role in the import of preproteins into mitochondria. J Biol Chem. 2005;280:7777–85.
Chacinska A, Lind M, Frazier AE, Dudek J, Meisinger C, Geissler A, et al. Mitochondrial presequence translocase: switching between TOM tethering and motor recruitment involves Tim21 and Tim17. Cell. 2005;120:817–29.
Hyman E, Kauraniemi P, Hautaniemi S, Wolf M, Mousses S, Rozenblum E, et al. Impact of DNA amplification on gene expression patterns in breast cancer. Cancer Res. 2002;62:6240–5.
Corson TW, Huang A, Tsao MS, Gallie BL. KIF14 is a candidate oncogene in the 1q minimal region of genomic gain in multiple cancers. Oncogene. 2005;24:4741–53.
Naylor TL, Greshock J, Wang Y, Colligon T, Yu QC, Clemmer V, et al. High resolution genomic analysis of sporadic breast cancer using array-based comparative genomic hybridization. Breast Cancer Res. 2005;7:R1186–98.
Jiang WG, Watkins G, Lane J, Cunnick GH, Douglas-Jones A, Mokbel K, et al. Prognostic value of rho GTPases and rho guanine nucleotide dissociation inhibitors in human breast cancers. Clin Cancer Res. 2003;9:6432–40.
Jiang WG, Douglas-Jones A, Mansel RE. Expression of peroxisome-proliferator activated receptor-gamma (PPARgamma) and the PPARgamma co-activator, PGC-1, in human breast cancer correlates with clinical outcomes. Int J Cancer. 2003;106:752–7.
Jiang WG, Watkins G, Fodstad O, Douglas-Jones A, Mokbel K, Mansel RE. Differential expression of the CCN family members Cyr61, CTGF and Nov in human breast cancer. Endocr Relat Cancer. 2004;11:781–91.
Wang C, Youle RJ. The role of mitochondria in apoptosis. Annu Rev Genet. 2009;43:95–118.
Wasilewski M, Scorrano L. The changing shape of mitochondrial apoptosis. Trends Endocrinol Metab. 2009;20:287–94.
Valko M, Rhodes CJ, Moncol J, Izakovic M, Mazur M. Free radicals, metals and antioxidants in oxidative stress-induced cancer. Chem Biol Interact. 2006;160:1–40.
Singh KK, Kulawiec M. Mitochondrial DNA polymorphism and risk of cancer. Methods Mol Biol. 2009;471:291–303.
Brandon M, Baldi P, Wallace DC. Mitochondrial mutations in cancer. Oncogene. 2006;25:4647–62.
Ma Y, Bai RK, Trieu R, Wong LJ. Mitochondrial dysfunction in human breast cancer cells and their transmitochondrial cybrids. Biochim Biophys Acta. 2010;1797:29–37.
Singh KK, Ayyasamy V, Owens KM, Koul MS, Vujcic M. Mutations in mitochondrial DNA polymerase-gamma promote breast tumorigenesis. J Hum Genet. 2009;54:516–24.
Kulawiec M, Owens KM, Singh KK. Cancer cell mitochondria confer apoptosis resistance and promote metastasis. Cancer Biol Ther. 2009;8:1378–85.
Radpour R, Fan AX, Kohler C, Holzgreve W, Zhong XY. Current understanding of mitochondrial DNA in breast cancer. Breast J. 2009;15:505–9.
Kannangai R, Vivekanandan P, Martinez-Murillo F, Choti M, Torbenson M. Fibrolamellar carcinomas show overexpression of genes in the RAS, MAPK, PIK3, and xenobiotic degradation pathways. Hum Pathol. 2007;38:639–44.
Xu X, Qiao M, Zhang Y, Jiang Y, Wei P, Yao J, et al. Quantitative proteomics study of breast cancer cell lines isolated from a single patient: discovery of TIMM17A as a marker for breast cancer. Proteomics. 2010;10:1374–90.
Iacovino M, Granycome C, Sembongi H, Bokori-Brown M, Butow RA, Holt IJ, et al. The conserved translocase Tim17 prevents mitochondrial DNA loss. Hum Mol Genet. 2009;18:65–74.
Shen J, Platek M, Mahasneh A, Ambrosone CB, Zhao H. Mitochondrial copy number and risk of breast cancer: a pilot study. Mitochondrion. 2010;10:62–8.
Mattingly KA, Ivanova MM, Riggs KA, Wickramasinghe NS, Barch MJ, Klinge CM. Estradiol stimulates transcription of nuclear respiratory factor-1 and increases mitochondrial biogenesis. Mol Endocrinol. 2008;22:609–22.
Chen JQ, Cammarata PR, Baines CP, Yager JD. Regulation of mitochondrial respiratory chain biogenesis by estrogens/estrogen receptors and physiological, pathological and pharmacological implications. Biochim Biophys Acta. 2009;1793:1540–70.
Chen JQ, Yager JD, Russo J. Regulation of mitochondrial respiratory chain structure and function by estrogens/estrogen receptors and potential physiological/pathophysiological implications. Biochim Biophys Acta. 2005;1746:1–17.
Kang D, Hamasaki N. Mitochondrial oxidative stress and mitochondrial DNA. Clin Chem Lab Med. 2003;41:1281–8.
Sastre-Serra J, Valle A, Company MM, Garau I, Oliver J, Roca P. Estrogen down-regulates uncoupling proteins and increases oxidative stress in breast cancer. Free Radic Biol Med. 2010;48:506–12.
Zhu W, Qin W, Bradley P, Wessel A, Puckett CL, Sauter ER. Mitochondrial DNA mutations in breast cancer tissue and in matched nipple aspirate fluid. Carcinogenesis. 2005;26:145–52.
Tseng LM, Yin PH, Chi CW, Hsu CY, Wu CW, Lee LM, et al. Mitochondrial DNA mutations and mitochondrial DNA depletion in breast cancer. Genes Chromosomes Cancer. 2006;45:629–38.
Erol A. Retrograde regulation due to mitochondrial dysfunction may be an important mechanism for carcinogenesis. Med Hypotheses. 2005;65:525–9.
Felty Q, Xiong WC, Sun D, Sarkar S, Singh KP, Parkash J, et al. Estrogen-induced mitochondrial reactive oxygen species as signal-transducing messengers. Biochemistry. 2005;44:6900–9.
Delsite R, Kachhap S, Anbazhagan R, Gabrielson E, Singh KK. Nuclear genes involved in mitochondria-to-nucleus communication in breast cancer cells. Mol Cancer. 2002;1:6.
Palmer HJ, Paulson KE. Reactive oxygen species and antioxidants in signal transduction and gene expression. Nutr Rev. 1997;55:353–61.
Thannickal VJ, Fanburg BL. Reactive oxygen species in cell signaling. Am J Physiol Lung Cell Mol Physiol. 2000;279:L1005–28.
Augenlicht LH, Heerdt BG. Mitochondria: integrators in tumorigenesis? Nat Genet. 2001;28:104–5.
Ishikawa K, Takenaga K, Akimoto M, Koshikawa N, Yamaguchi A, Imanishi H, et al. ROS-generating mitochondrial DNA mutations can regulate tumor cell metastasis. Science. 2008;320:661–4.
Conflict of interest
None.
Author information
Authors and Affiliations
Corresponding author
About this article
Cite this article
Salhab, M., Patani, N., Jiang, W. et al. High TIMM17A expression is associated with adverse pathological and clinical outcomes in human breast cancer. Breast Cancer 19, 153–160 (2012). https://doi.org/10.1007/s12282-010-0228-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12282-010-0228-3