Abstract
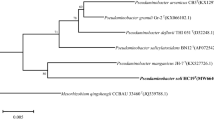
A Gram negative, aerobic, non-motile and rod-shaped bacterial strain designated as Gr-2T was isolated from granules used in a wastewater treatment plant in Korea, and its taxonomic position was investigated using a polyphasic approach. Strain Gr-2T grew at 18–37°C (optimum temperature, 30°C) and a pH of 6.0–8.0 (optimum pH, 7.0) on R2A agar medium. Based on 16S rRNA gene phylogeny, the novel strain showed a new branch within the genus Pseudaminobacter of the family Phyllobacteriaceae, and formed clusters with Pseudaminobacter defluvii THI 051T (98.9%) and Pseudaminobacter salicylatoxidans BN12T (98.7%). The G+C content of the genomic DNA was 63.6%. The predominant respiratory quinone was ubiquinone-10 (Q-10) and the major fatty acids were cyclo-C19:0 ω8c, C18:1 ω7c, and iso-C17:0. The overall polar lipid patterns of Gr-2T were similar to those determined for the other Pseudaminobacter species. DNA-DNA relatedness values between strain Gr-2T and its closest phylogenetically neighbors were below 18%. Strain Gr-2T could be differentiated genotypically and phenotypically from the recognized species of the genus Pseudaminobacter. The isolate therefore represents a novel species, for which the name Pseudaminobacter granuli sp. nov. is proposed with the type strain Gr-2T (=KACC 18877T =LMG 29567T).
Similar content being viewed by others
References
Atlas, R.M. 1993. Handbook of Microbiological Media. CRC Press, Boca Raton, Florida, USA.
Buck, J.D. 1982. Nonstaining (KOH) method for determination of Gram reactions of marine bacteria. Appl. Environ. Microbiol. 44, 992–993.
Cappuccino, J.G. and Sherman, N. 2002. Microbiology: a laboratory manual, 6th ed. Pearson Education, Inc., California, USA.
Ezaki, T., Hashimoto, Y., and Yabuuchi, E. 1989. Fluorometric deoxyribonucleic acid-deoxyribonucleic acid hybridization in microdilution wells as an alternative to membrane filter hybridization in which radioisotopes are used to determine genetic relatedness among bacterial strains. Int. J. Syst. Bacteriol. 39, 224–229.
Felsenstein, J. 1985. Confidence limit on phylogenies: an approach using the bootstrap. Evolution 39, 783–791.
Fitch, W.M. 1971. Toward defining the course of evolution: minimum change for a specific tree topology. Syst. Zool. 20, 406–416.
Hall, T.A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98.
Hiraishi, A., Ueda, Y., Ishihara, J., and Mori, T. 1996. Comparative lipoquinone analysis of influent sewage and activated sludge by high-performance liquid chromatography and photodiode array detection. J. gen. Appl. Microbiol. 42, 457–469.
Kämpfer, P., Müller, C., Mau, M., Neef, A., Auling, G., Busse, H.J., Osborn, A.M., and Stolz, A. 1999. Description of Pseudaminobacter gen. nov. with two new species, Pseudaminobacter salicylatoxidans sp. nov. and Pseudaminobacter defluvii sp. nov. Int. J. Syst. Bacteriol. 49, 887–897.
Kim, J.K., Kang, M.S., Park, S.C., Kim, K.M., Choi, K., Yoon, M.H., and Im, W.T. 2015. Sphingosinicella ginsenosidimutans sp. nov., with ginsenoside converting activity. J. Microbiol. 53, 435–441.
Kimura, M. 1983. The Neutral Theory of Molecular Evolution. Cambridge: Cambridge University Press, Cambridge, New York, USA.
Mesbah, M., Premachandran, U., and Whitman, W. 1989. Precise measurement of the G+C content of deoxyribonucleic acid by high performance liquid chromatography. Int. J. Syst. Bacteriol. 39, 159–167.
Minnikin, D.E., O’Donnell, A.G., Goodfellow, M., Alderson, G., Athalye, M., Schaal, A., and Parlett, J.H. 1984. An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J. Microbiol. Methods 2, 233–241.
Moore, D.D. and Dowhan, D. 1995. Preparation and analysis of DNA, pp. 2–11. In Ausubel, F.W., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., and Struhl, K. (eds.), Current Protocols in Molecular Biology. Wiley, New York, USA.
Perry, L.B. 1973. Gliding motility in some non-spreading flexibacteria. J. Appl. Bacteriol. 36, 227–232.
Saitou, N. and Nei, M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425.
Sasser, M. 1990. Identification of bacteria by gas chromatography of cellular fatty acids. MIDI Technical Note 101. MIDI Inc., Newark, DE,USA.
Schenkel, E., Berlaimont, V., Dubois, J., Helson-Cambier, M., and Hanocq, M. 1995. Improved high-performance liquid chromatographic method for the determination of polyamines as their benzoylated derivatives: application to P388 cancer cells. J. Chromatogr. B. 668, 189–197.
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 30, 2725–2729.
Ten, L.N., Im, W.T., Kim, M.K., Kang, M.S., and Lee, S.T. 2004. Development of a plate technique for screening of polysaccharide-degrading microorganisms by using a mixture of insoluble chromogenic substrates. J. Microbiol. Methods 56, 375–382.
Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., and Higgins, D.G. 1997. The Clustal_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24, 4876–4882.
Wayne, L.G., Brenner, D.J., Colwell, R.R., Grimont, P.A.D., Kandler, O., Krichevsky, M.I., Moore, L.H., Moore, W.E.C., Murray, R.G.E., Stackebrandt, E., et al. 1987. International Committee on Systematic Bacteriology. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int. J. Syst. Bacteriol. 37, 463–464.
Yoon, S.H., Ha, S.M., Kwon, S., Lim, J., Kim, Y., Seo, H., and Chun, J. 2017. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole genome assemblies. Int. J. Syst. Evol. Microbiol. 67, 1613–1617.
Author information
Authors and Affiliations
Corresponding authors
Additional information
The GenBank accession number for the 16S rRNA gene sequence of strain Gr-2T is KX066102.
Rights and permissions
About this article
Cite this article
Hahn, Y.K., Kim, M.S. & Im, WT. Pseudaminobacter granuli sp. nov., isolated from granules used in a wastewater treatment plant. J Microbiol. 55, 607–611 (2017). https://doi.org/10.1007/s12275-017-7257-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12275-017-7257-y