Abstract
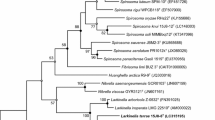
A Gram-negative, short, rod-shaped bacterium, TSA1T, was isolated from forest soil collected at Kyonggi University, South Korea. Assessment of 16S rRNA gene sequence similarity indicated that the strain is related to Massilia niastensis 5516S-1T (98.3%), M. haematophila CCUG 38318T (97.9%), M. aerilata 5516S-11T (97.9%), M. tieshanensis TS3T (97.6%), and M. varians CCUG 3529T (97.1%). Colonies grown on Reasoner’s 2A agar at 30°C for 2 days were transparent, white, round, smooth, and glossy. The cells grew at 10–42°C (optimum: 25–37°C) and pH 5–9 (optimum: 5–9) and in 0–2% NaCl (optimum: 0–1%). TSA1T was able to grow on trypticase soy and nutrient agar, but not on Luria-Bertani or MacConkey agar. The strain was catalase- and oxidasepositive and able to degrade starch and casein, but not carboxymethyl cellulose. The predominant quinone of TSA1T was Q-8, the major fatty acids were summed feature 3 and C16:0, and the DNA G+C content was 66.7 mol%. Given these findings, we propose that this strain is a novel species of the genus Massilia. We suggest the name Massilia kyonggiensis sp. nov. (type strain, KACC 17471T =KEMB 9005-031T =JCM 19189T).
Similar content being viewed by others
References
Collins, M.D. and Jones, D. 1981. Distribution of isoprenoid quinone structural types in bacteria and their taxonomic implications. Microbiol. Rev. 45, 316–354.
Doetsch, R.N. 1981. Determinative methods of light microscopy, pp. 21–33. In Gerhardt, P., Murray, R.G.E., Costilow, R.N., Nester, E.W., Wood, W.A., Krieg, N.R., and Phillips, G. (eds.), Manual of Methods for General Bacteriology. American Society for Microbiology, Washington, D.C., USA.
Du, Y., Yu, X., and Wang, G. 2012. Massilia tieshanensis sp. nov., isolated from mining soil. Int. J. Syst. Evol. Microbiol. 62, 2356–2362.
Felsenstein, J. 1985. Confidence limit on phylogenies: an approach using the bootstrap. Evolution 39, 783–791.
Hall, T.A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98.
Johnson, T.R. and Case, C.L. 2007. Laboratory Experiments in Microbiology (8th ed.), pp. 141–143. Pearson Education, Inc., San Francisco, California, USA.
Kim, O.S., Cho, Y.J., Lee, K., Yoon, S.H., Kim, M., Na, H., Park, S.C., Jeon, Y.S., Lee, J.H., Yi, H., and et al. 2012. Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int. J. Syst. Evol. Microbiol. 62, 716–721.
Komagata, K. and Suzuki, K. 1987. Lipid and cell-wall analysis in bacterial systematics. Methods Microbiol. 19, 161–207.
La Scola, B., Birtles, R.J., Mallet, M.N., and Raoult, D. 1998. Massilia timonae gen. nov., sp. nov., isolated from blood of an immunocompromised patient with cerebellar lesions. J. Clin. Microbiol. 36, 2847–2852.
Mehlen, A., Goeldner, M., Ried, S., Stindl, S., Ludwig, W., and Schleifer, K.H. 2004. Development of a fast DNA-DNA hybridization method based on melting profiles in microplates. Syst. Appl. Microbiol. 27, 689–695.
Mesbah, M., Premachandran, U., and Whitman, W.B. 1989. Precise measurement of the G+C content of deoxyribonucleic acid by high-performance liquid chromatography. Int. J. Syst. Bacteriol. 39, 159–167.
Minnikin, D.E., O’Donnell, A.G., Goodfellow, M., Alderson, G., Athalye, M., Schaal, A., and Parlett, J.H. 1984. An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J. Microbiol. Methods 2, 233–241.
Pham, T.H.V. and Kim, J. 2012. Cultivation of unculturable soil bacteria. Trends Biotechnol. 30, 475–484.
Pham, T.H.V. and Kim, J. 2013. Bacillus thaonhiensis sp. nov., a new species, was isolated from the forest soil of Kyonggi University by using a modified culture method. Curr. Microbiol. 68, 88–95.
Saitou, N. and Nei, M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425.
Sasser, M. 1990. Identification of bacteria by gas chromatography of cellular fatty acids. In MIDI Technical Note 101. MIDI Inc., Newark, DE, USA.
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739.
Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., and Higgins, D.G. 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24, 4876–4882.
Wang, J.W., Zhang, J.L., Pang, H.C., Zhang, Y.B., Li, Y.Y., and Fan, J.P. 2012. Massilia flava sp. nov., isolated from soil. Int. J. Syst. Evol. Microbiol. 62, 580–585.
Zhang, Y.Q., Li, W.J., Zhang, K.Y., Tian, X.P., Jiang, Y., Xu, L.H., Jiang, C.L., and Lai, R. 2006. Massilia dura sp. nov., Massilia albidiflava sp. nov., Massilia plicata sp. nov. and Massilia lutea sp. nov., isolated from soils in China. Int. J. Syst. Evol. Microbiol. 56, 459–463.
Zul, D., Wanner, G., and Overmann, J. 2008. Massilia brevitalea sp. nov., a novel betaproteobacterium isolated from lysimeter soil. Int. J. Syst. Evol. Microbiol. 58, 1245–1251.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kim, J. Massilia kyonggiensis sp. nov., isolated from forest soil in Korea. J Microbiol. 52, 378–383 (2014). https://doi.org/10.1007/s12275-014-4010-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12275-014-4010-7