Abstract
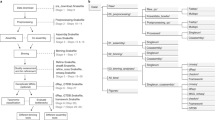
Metagenomic and metatranscriptomic shotgun sequencing techniques are gaining popularity as more cost-effective next-generation sequencing technologies become commercially available. The initial stage of bioinfor-matic analysis generally involves the identification of phylogenetic markers such as ribosomal RNA genes. The sequencing reads that do not code for rRNA can then be used for protein-based analysis. Hidden Markov model is a well-known method for pattern recognition. Hidden Markov models that are trained on well-curated rRNA sequence databases have been successfully used to identify DNA sequence coding for rRNAs in pro-karyotes. Here, we introduce rRNASelector, which is a computer program for selecting rRNA genes from massive metagenomic and metatranscriptomic sequences using hidden Markov models. The program successfully identified prokaryotic 5S, 26S, and 23S rRNA genes from Roche 454 FLX Titanium-based metagenomic and metatranscriptomic libraries. The rRNASelector program is available at http://sw.ezbiocloud.net/rrnaselector.
Similar content being viewed by others
References
Chun, J., J.H. Lee, Y. Jung, M. Kim, S. Kim, B.K. Kim, and Y.W. Lim. 2007. Eztaxon: A web-based tool for the identification of prokaryotes based on 16S ribosomal rna gene sequences. Int. J. Syst. Evol. Microbiol. 57, 2259–2261.
Eddy, S.R. 1998. Profile hidden markov models. Bioinformatics 14, 755–763.
Eddy, S.R. 2009. A new generation of homology search tools based on probabilistic inference. Genome Inform. 23, 205–211.
Glass, E.M., J. Wilkening, A. Wilke, D. Antonopoulos, and F. Meyer. 2010. Using the metagenomics rast server (mg-rast) for analyzing shotgun metagenomes. Cold Spring Harb. Protoc. 2010, pdb prot5368.
Handelsman, J. 2004. Metagenomics: Application of genomics to uncultured microorganisms. Microbiol. Mol. Biol. Rev. 68, 669–685.
Handelsman, J., M.R. Rondon, S.F. Brady, J. Clardy, and R.M. Goodman. 1998. Molecular biological access to the chemistry of unknown soil microbes: A new frontier for natural products. Chem. Biol. 5, R245–249.
Huang, Y., P. Gilna, and W. Li. 2009. Identification of ribosomal RNA genes in metagenomic fragments. Bioinformatics 25, 1338–1340.
Huson, D.H., A.F. Auch, J. Qi, and S.C. Schuster. 2007. Megan analysis of metagenomic data. Genome Res. 17, 377–386.
Lagesen, K., P. Hallin, E.A. Rodland, H.H. Staerfeldt, T. Rognes, and D.W. Ussery. 2007. Rnammer: Consistent and rapid annotation of ribosomal rna genes. Nucleic Acids Res. 35, 3100–3108.
Mardis, E.R. 2008. Next-generation DNA sequencing methods. Annu. Rev. Genomics Hum. Genet. 9, 387–402.
Margulies, M., M. Egholm, W.E. Altman, S. Attiya, J.S. Bader, L.A. Bemben, J. Berka, and et al. 2005. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437, 376–380.
Meyer, F., D. Paarmann, M. D-Souza, R. Olson, E.M. Glass, M. Kubal, T. Paczian, and et al. 2008. The metagenomics rast server — a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics 9, 386.
Polz, M.F. and C.M. Cavanaugh. 1998. Bias in template-to-product ratios in multitemplate PCR. Appl. Environ. Microbiol. 64, 3724–3730.
Qin, J., R. Li, J. Raes, M. Arumugam, K.S. Burgdorf, C. Manichanh, T. Nielsen, and et al. 2010. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65.
Rondon, M.R., P.R. August, A.D. Bettermann, S.F. Brady, T.H. Grossman, M.R. Liles, K.A. Loiacono, and et al. 2000. Cloning the soil metagenome: A strategy for accessing the genetic and functional diversity of uncultured microorganisms. Appl. Environ. Microbiol. 66, 2541–2547.
Schmidt, T.M., E.F. DeLong, and N.R. Pace. 1991. Analysis of a marine picoplankton community by 16S rRNA gene cloning and sequencing. J. Bacteriol. 173, 4371–4378.
Szymanski, M., M.Z. Barciszewska, V.A. Erdmann, and J. Barciszewski. 2002. 5S ribosomal RNA database. Nucleic Acids Res. 30, 176–178.
Valouev, A., J. Ichikawa, T. Tonthat, J. Stuart, S. Ranade, H. Peckham, K. Zeng, and et al. 2008. A high-resolution, nucleosome position map of c. Elegans reveals a lack of universal sequence-dictated positioning. Genome Res. 18, 1051–1063.
Venter, J.C., K. Remington, J.F. Heidelberg, A.L. Halpern, D. Rusch, J.A. Eisen, D. Wu, and et al. 2004. Environmental genome shotgun sequencing of the sargasso sea. Science 304, 66–74.
von Wintzingerode, F., U.B. Gobel, and E. Stackebrandt. 1997. Determination of microbial diversity in environmental samples: Pitfalls of PCR-based rRNA analysis. FEMS Microbiol. Rev. 21, 213–229.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Lee, JH., Yi, H. & Chun, J. rRNASelector: A computer program for selecting ribosomal RNA encoding sequences from metagenomic and metatranscriptomic shotgun libraries. J Microbiol. 49, 689–691 (2011). https://doi.org/10.1007/s12275-011-1213-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12275-011-1213-z