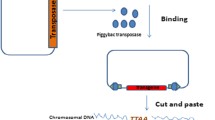
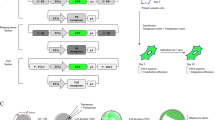
Abstract
Transposons are widely used for genetic engineering in various model organisms. Recently, piggyBac (PB) has been developed as a transposable and efficient gene transfer tool in mammalian cells. In the present study, we developed three types of PB transposon systems containing a dual plasmid system (DPS), a single plasmid system (SPS), and a DNA-mRNA combined system (DRPS) and characterized their basic properties in HEK293 cells. The basic elements of the donor plasmid included a selectable-reporter gene expression cassette, two loxP sites in the same orientation, a multiple cloning site, and two chicken β-globin insulator core elements. We further identified the function of the selectable-reporter and examined PB integration sites in the human genome. Moreover, we compared the transposition efficacy and found that SPS transposed more efficiently, as compared to DPS; integration into the host genome was determined by measuring PBase activity. Results discovered the loss of PBase activity in the DRPS, indicating that this system is much more biologically safe, as compared to DPS and SPS. Finally, we employed the DRPS to successfully perform a gene delivery into bovine mammary epithelial cells (BMECs). Taken together, the information from this study will improve the flexibility of PB transposon systems and reduce the genotoxicity of PBase in genetic engineering.
Similar content being viewed by others
References
Lacoste, A., F. Berenshteyn, and A. H. Brivanlou (2009) An efficient and reversible transposable system for gene delivery and lineage-specific differentiation in human embryonic stem cells. Cell Stem Cell 5: 332–342.
Cary, L. C., M. Goebel, B. G. Corsaro, H. G. Wang, E. Rosen, and M. J. Fraser (1989) Transposon mutagenesis of baculoviruses: Analysis of Trichoplusia ni transposon IFP2 insertions within the FP-locus of nuclear polyhedrosis viruses. Virol. 172: 156–169.
Fraser, M. J., L. Cary, K. Boonvisudhi, and H. G. H. Wang (1995) Assay for Movement of lepidopteran transposon Ifp2 in insect cells using a baculovirus genome as a target DNA. Virol. 211: 397–407.
Fraser, M. J., T. Coszczon, T. Elick, and C. Bauser (1996) Precise excision of TTAA-specific lepidopteran transposons piggy-Bac(IFP2) and tagalong (TFP3) from the baculovirus genome in cell lines from two species of Lepidoptera. Insect Mol. Biol. 5: 141–151.
Handler, A. M., S. D. McCombs, M. J. Fraser, and S. H. Saul (1998) The lepidopteran transposon vector, piggyBac, mediates germ-line transformation in the Mediterranean fruit fly. Proc. Nat. Acad. Sci. U. S. A. 95: 7520–7525.
Lynch, A. G., F. Tanzer, M. J. Fraser, E. G. Shephard, A. -L. Williamson, and E. P. Rybicki (2010) Use of the piggyBac transposon to create HIV-1 gag transgenic insect cell lines for continuous VLP production. BMC Biotechnol. 10: 30.
Wilson, M. H., C. J. Coates, and A. L. George (2007) piggyBac transposon-mediated gene transfer in human cells. Mol. Therapy: The J. American Soc. Gene Therapy 15: 139–145.
Ding, S., X. Wu, G. Li, M. Han, Y. Zhuang, and T. Xu (2005) Efficient transposition of the piggyBac (PB) transposon in mammalian cells and mice. Cell. 122: 473–483.
Rad, R., L. Rad, W. Wang, J. Cadinanos, G. Vassiliou, S. Rice, L. S. Campos, K. Yusa, R. Banerjee, M. A. Li, J. de la Rosa, A. Strong, D. Lu, P. Ellis, N. Conte, F. T. Yang, P. Liu, and A. Bradley (2010) piggyBac transposon mutagenesis: A tool for cancer gene discovery in mice. Sci. 330: 1104–1107.
Chew, S. K., R. Rad, P. A. Futreal, A. Bradley, and P. T. Liu (2011) Genetic screens using the piggyBac transposon. Methods 53: 366–371.
Jang, G., S. Kim, S. Islam, W. Choi, S. Lee, W. Lee, B. Lee, J. Cho, and J. Moon (2011) Production of transgenic bovine cloned embryos using piggyBac transposition. Transg. Res. 20: 1176–1177.
Bai, D. P., M. M. Yang, and Y. L. Chen (2012) piggyBac transposon-mediated gene transfer in Cashmere goat fetal fibroblast cells. Biosci. Biotechnol. Biochem. 76: 933–937.
Park, T. S. and J. Y. Han (2012) piggyBac transposition into primordial germ cells is an efficient tool for transgenesis in chickens. Proc. Nat. Acad. Sci. U. S. A. 109: 9337–9341.
Choi, W. J., S. J. Lee, W. W. Lee, S. J. Kim, I. M. Saadeldin, J. K. Cho, B. C. Lee, and G. Jang (2013) Implantation of transgenic bovine cloned embryos derived from transfected cells by piggybac transposition. Reprod. Fert. Develop. 25: 173–174.
Kim, S. J., I. M. Saadeldin, W. J. Choi, S. J. Lee, W. W. Lee, B. H. Kim, H. J. Han, D. H. Bang, B. C. Lee, and G. Jang (2011) Production of transgenic bovine cloned embryos using piggybac transposition. J. Veterinary Med. Sci. 73: 1453–1457.
Woltjen, K., I. P. Michael, P. Mohseni, Desai R, M. Mileikovsky, R. Hamalainen, R. Cowling, W. Wang, P. Liu, M. Gertsenstein, K. Kaji, H. K. Sung, and A. Nagy (2009) piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature 458: 766–770.
Yusa, K., R. Rad, J. Takeda, and A. Bradley (2009) Generation of transgene-free induced pluripotent mouse stem cells by the piggyBac transposon. Nat. Methods 6: 363–369.
Tsukiyama, T. (2011) Development of a simple and efficient method for generation of iPS cells by using piggybac system to screen novel culture conditions. Seikagaku The J. Japan. Biochem. Soc. 83: 855–858.
Tsukiyama, T., R. Asano, T. Kawaguchi, N. Kim, M. Yamada, N. Minami, Y. Ohinata, and H. Imai (2011) Simple and efficient method for generation of induced pluripotent stem cells using piggyBac transposition of doxycycline-inducible factors and an EOS reporter system. Genes to Cells: Devot. Mol. Cell. Mech. 16: 815–825.
Zhou, F., S. Liang, A. H. Chen, C. O. Singh, R. Bhaskar, Y. S. Niu, and Y. G. Miao (2012) A transgenic Marc-145 cell line of piggyBac transposon-derived targeting shRNA interference against porcine reproductive and respiratory syndrome virus. Vet. Res. Commun. 36: 99–105.
Uetake, H., K. Oka, and Y. Niki (2011) Stable transformation and cloning mediated by piggyBac vector and RNA interference knockdown of Drosophila ovarian cell line. In vitro Cell. Develop. Biol. Animal. 47: 689–694.
Yusa, K., L. Zhou, M. A. Li, A. Bradley, and N. L. Craig (2011) A hyperactive piggyBac transposase for mammalian applications. Proc. Nat. Acad. Sci. U. S. A. 108: 1531–1536.
Chen, Y. T., K. Furushima, P. S. Hou, A. T. Ku, J. M. Deng, C. W. Jang, H. Fang, H. P. Adams, M. L. Kuo, H. N. Ho, C. L. Chien, and R. R. Behringer (2010) piggyBac transposon-mediated, reversible gene transfer in human embryonic stem cells. Stem Cells and Develop. 19: 763–771.
Urschitz, J., M. Kawasumi, J. Owens, K. Morozumi, H. Yamashiro, I. Stoytchev, J. Marh, J. A. Dee, K. Kawamoto, C. J. Coates, J. M. Kaminski, P. Pelczar, R. Yanagimachi, and S. Moisyadi (2010) Helper-independent piggyBac plasmids for gene delivery approaches: strategies for avoiding potential genotoxic effects. Proc. Nat. Acad. Sci. U. S. A. 107: 8117–8122.
Li, X., R. A. Harrell, A. M. Handler, T. Beam, K. Hennessy, and M. J. Fraser (2005) piggyBac internal sequences are necessary for efficient transformation of target genomes. Insect Mol. Biol. 14: 17–30.
Krieg, P. and D. Melton (1984) Functional messenger RNAs are produced by SP6 in vitro transcription of cloned cDNAs. Nucleic Acids Res. 12: 7057–7070.
Warren, L., P. D. Manos, T. Ahfeldt, Y. H. Loh, H. Li, F. Lau, W. Ebina, P. K. Mandal, Z. D. Smith, A. Meissner, G. Q. Daley, A. S. Brack, J. J. Collins, C. Cowan, T. M. Schlaeger, and D. J. Rossi (2010) Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell. 7: 618–630.
Liu, Y. G. and R. F. Whittier (1995) Thermal asymmetric interlaced PCR: automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking. Genom. 25: 674–681.
Liu, Y. G. and Y. Chen (2007) High-efficiency thermal asymmetric interlaced PCR for amplification of unknown flanking sequences. BioTechniques. 43: 649–650.
Su, H., X. Liu, W. Yan, T. Shi, X. Zhao, D. P. Blake, F. M. Tomley, and X. Suo (2012) piggyBac transposon-mediated transgenesis in the apicomplexan parasite Eimeria tenella. PloS one. 7: e40075.
Nakanishi, H., Y. Higuchi, S. Kawakami, F. Yamashita, and M. Hashida (2011) Comparison of piggyBac transposition efficiency between linear and circular donor vectors in mammalian cells. J. Biotechnol. 154: 205–208.
Zhao, M. T., H. Lin, F. J. Liu, F. S. Quan, G. H. Wang, J. Liu, S. Hua, and Y. Zhang (2009) Efficiency of human lactoferrin transgenic donor cell preparation for SCNT. Theriogenol. 71: 376–384.
Wu, S. C., Y. J. Meir, C. J. Coates, A. M. Handler, P. Pelczar, S. Moisyadi, and J. M. Kaminski (2006) piggyBac is a flexible and highly active transposon as compared to sleeping beauty, Tol2, and Mos1 in mammalian cells. Proc. Nat. Acad. Sci. U. S. A. 103: 15008–15013.
Meir, Y. J., M. T. Weirauch, H. S. Yang, P. C. Chung, R. K. Yu, and S. C. Wu (2011) Genome-wide target profiling of piggyBac and Tol2 in HEK 293: Pros and cons for gene discovery and gene therapy. BMC Biotechnol. 11: 28.
Li, M. A., D. J. Turner, Z. Ning, K. Yusa, Q. Liang, S. Eckert, L. Rad, T. W. Fitzgerald, N. L. Craig, and A. Bradley (2011) Mobilization of giant piggyBac transposons in the mouse genome. Nucleic Acids Res. 39: e148.
Rostovskaya, M., J. Fu, M. Obst, I. Baer, S. Weidlich, H. Wang, A. J. Smith, K. Anastassiadis, and A. F. Stewart (2012) Transposon-mediated BAC transgenesis in human ES cells. Nucleic Acids Res. 40: e150.
Bire, S., D. Gosset, G. Jégot, P. Midoux, C. Pichon, and F. Rouleux-Bonnin (2013) Exogenous mRNA delivery and bioavailability in gene transfer mediated by piggyBac transposition. BMC Biotechnol. 13: 75.
Morales, M. E., V. H. Mann, K. J. Kines, G. N. Gobert, M. J. Fraser, B. H. Kalinna, J. M. Correnti, E. J. Pearce, and P. J. Brindley (2007) piggyBac transposon mediated transgenesis of the human blood fluke, Schistosoma mansoni. The FASEB J. 21: 3479–3489.
Shinmyo, Y., T. Mito, T. Matsushita, I. Sarashina, K. Miyawaki, H. Ohuchi, and S. Noji (2004) piggyBac-mediated somatic transformation of the two-spotted cricket, Gryllus bimaculatus. Develop. Growth & Diff. 46: 343–349.
Bire, S., D. Ley, S. Casteret, N. Mermod, Y. Bigot, and F. Rouleux-Bonnin (2013) Optimization of the piggyBac transposon using mRNA and Insulators: Toward a more reliable gene delivery system. PloS one. 8: e82559.
Maury, J. J., A. B. Choo, and K. K. Chan (2011) Technical advances to genetically engineering human embryonic stem cells. Integ. Biol: Quantitative Biosci. Nano to Macro. 3: 717–723.
Sarkar, A., C. Sim, Y. S. Hong, J. R. Hogan, M. J. Fraser, H. M. Robertson, and F. H. Collins (2003) Molecular evolutionary analysis of the widespread piggyBac transposon family and related “domesticated” sequences. Mol. Gen. Genom.: MGG. 270: 173–180.
Rivella, S., J. A. Callegari, C. May, C. W. Tan, and M. Sadelain (2000) The cHS4 insulator increases the probability of retroviral expression at random chromosomal integration sites. J. Virol. 74: 4679–4687.
Arumugam, P. I., J. Scholes, N. Perelman, P. Xia, J. K. Yee, and P. Malik (2007) Improved human beta-globin expression from self-inactivating lentiviral vectors carrying the chicken hypersensitive site-4 (cHS4) insulator element. Mol. Therapy 15: 1863–1871.
Li, C. L. and D. W. Emery (2008) The cHS4 chromatin insulator reduces gammaretroviral vector silencing by epigenetic modifications of integrated provirus. Gene Therapy 15: 49–53.
Sharma, N., A. K. Hollensen, R. O. Bak, N. H. Staunstrup, L. D. Schroder, and J. G. Mikkelsen (2012) The impact of cHS4 insulators on DNA transposon vector mobilization and silencing in retinal pigment epithelium cells. PloS one. 7: e48421.
Palmenberg, A. C., G. D. Parks, D. J. Hall, R. H. Ingraham, T. W. Seng, and P. V. Pallai (1992) Proteolytic processing of the cardioviral P2 region: Primary 2A/2B cleavage in clone-derived precursors. Virol. 190: 754–762.
Ryan, M. D., A. King, and G. P. Thomas (1991) Cleavage of footand-mouth disease virus polyprotein is mediated by residues located within a 19 amino acid sequence. The J. Gen. Virol. 72: 2727–2732.
Chinnasamy, D., M. D. Milsom, J. Shaffer, J. Neuenfeldt, A. F. Shaaban, G. P. Margison, L. J. Fairbairn, and N. Chinnasamy (2006) Multicistronic lentiviral vectors containing the FMDV 2A cleavage factor demonstrate robust expression of encoded genes at limiting MOI. Virol J. 3: 14.
Schwenk, F., U. Baron, and K. Rajewsky (1995) A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic Acids Res. 23: 5080–5081.
Betz, U. A., C. A. Vosshenrich, K. Rajewsky, and W. Muller (1996) Bypass of lethality with mosaic mice generated by CreloxP-mediated recombination. Curr. Biol.: CB. 6: 1307–1316.
Ruby, K. M. and B. Zheng (2009) Gene targeting in a HUES line of human embryonic stem cells via electroporation. Stem Cells. 27: 1496–1506.
Yu, Y., Y. Wang, Q. Tong, X. Liu, F. Su, F. Quan, Z. Guo, and Y. Zhang (2013) A site-specific recombinase-based method to produce antibiotic selectable marker free transgenic cattle. PloS one. 8: e62457.
Clark, A. J. (1998) The mammary gland as a bioreactor: Expression, processing, and production of recombinant proteins. J. Mammary Gland Biol. 3: 337–350.
Yang, P. H., J. W. Wang, G. C. Gong, X. Z. Sun, R. Zhang, Z. Du, Y. Liu, R. Li, F. R. Ding, B. Tang, Y. P. Dai, and N. Li (2008) Cattle mammary bioreactor generated by a novel procedure of transgenic cloning for large-scale production of functional human lactoferrin. PloS one. 3.
Zakhartchenko, V., R. Alberio, M. Stojkovic, K. Prelle, W. Schernthaner, P. Stojkovic, H. Wenigerkind, R. Wanke, M. Duchler, R. Steinborn, M. Mueller, G. Brem, and E. Wolf (1999) Adult cloning in cattle: Potential of nuclei from a permanent cell line and from primary cultures. Mol. Reproduc. Develop. 54: 264–272.
Kishi, M, Y. Itagaki, R. Takakura, M. Imamura, T. Sudo, M. Yoshinari, M. Tanimoto, H. Yasue, and N. Kashima (2000) Nuclear transfer in cattle using colostrum-derived mammary gland epithelial cells and ear-derived fibroblast cells. Theriogenol. 54: 675–684.
Kishi M., Y. Itagaki, T. Sudo, and R. Takakura (2003) In vitro development of bovine nuclear transfer embryos reconstructed with mammary gland epithelial cells at different passages. Animal Sci. J. 74: 363–368.
Akagi, S., S. Takahashi, K. Ohkoshi, T. Takenouchi, M. Shimizu, M. Geshi, N. Adachi, D. Fuchimoto, Y. Izaike, and H. Aso (2002) Nuclear transfer using a bovine mammary epithelial cell line (BMEC). Animal Sci. J. 73: 465–469.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Hu, G., Wang, J., Huang, H. et al. A modified piggybac transposon system mediated by exogenous mRNA to perform gene delivery in bovine mammary epithelial cells. Biotechnol Bioproc E 19, 350–362 (2014). https://doi.org/10.1007/s12257-013-0811-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12257-013-0811-5