Abstract
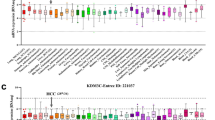
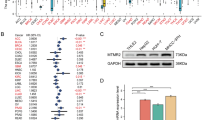
The multiple myeloma SET domain (MMSET) involved in the t(4;14)(p16;q32) chromosomal translocation encodes a histone lysine methyltransferase. High expression of MMSET is common translocation in multiple myeloma (MM) and is associated with the worst prognosis. Recent studies have shown that overexpression of MMSET is significant in other tumor types compared to their normal tissues. However, little is known about its role in hepatocellular carcinoma (HCC). In these study we investigate the expression of MMSET in HCC and to make correlations with clinicopathologic features. Twenty-eight pairs of HCC and adjacent non-tumor tissues, and eight normal liver tissues were collected for MMSET detection by western blotting and real time-PCR analysis. Immunohistochemistry was used to determine the expression of MMSET in HCC and adjacent non-tumor tissues from 103 patients. Overexpression of MMSET was significantly associated with Edmondson stage, vascular invasion. Moreover, Kaplan-Meier curves showed that MMSET upregulated was associated with shorter overall survival and disease-free survival in HCC patient. In conclusion, our study demonstrates for the first time that overexpression of MMSET is an independent prognostic factor and is correlated with poor survival in HCC patients.



Similar content being viewed by others
References
Di Bisceglie AM (2004) Issues in screening and surveillance for hepatocellular carcinoma. Gastroenterology 127:S104–S107
Szabó E, Páska C, Kaposi Novák P, Schaff Z, Kiss A (2004) Similarities and differences in hepatitis B and C virus induced hepatocarcinogenesis. Pathol Oncol Res 10(1):5–11
El-Serag HB, Rudolph KL (2007) Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology 132(7):2557–2576
Xie B, Wang DH, Spechler SJ (2012) Sorafenib for treatment of hepatocellular carcinoma: a systematic review. Dig Dis Sci 57(5):1122–1129
Martin C, Zhang Y (2005) The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol 6(11):838–849
Li Y, Trojer P, Xu CF et al (2009) The target of the NSD family of histone lysine methyltransferases depends on the nature of the substrate. J Biol Chem 284(49):34283–34295
Stec I, Wright TJ, van Ommen GJ et al (1998) WHSC1, a 90 kb SET domain-containing gene, expressed in early development and homologous to a Drosophila dysmorphy gene maps in the Wolf-Hirschhorn syndrome critical region and is fused to IgH in t(4;14) multiple myeloma. Hum Mol Genet 7(7):1071–1082
Chesi M, Nardini E, Lim RS, Smith KD, Kuehl WM, Bergsagel PL (1998) The t(4;14) translocation in myeloma dysregulates both FGFR3 and a novel gene, MMSET, resulting in IgH/MMSET hybrid transcripts. Blood 92(9):3025–3034
Keats JJ, Reiman T, Belch AR, Pilarski LM (2006) Ten years and counting: so what do we know about t(4;14)(p16;q32) multiple myeloma. Leuk Lymphoma 47(11):2289–2300
Hudlebusch HR, Skotte J, Santoni-Rugiu E et al (2011) MMSET is highly expressed and associated with aggressiveness in neuroblastoma. Cancer Res 71(12):4226–4235
Li J, Yin C, Okamoto H, Mushlin H et al (2008) Identification of a novel proliferation-related protein, WHSC1 4a, in human gliomas. Neuro Oncol 10(1):45–51
Kang HB, Choi Y, Lee JM et al (2009) The histone methyltransferase, NSD2, enhances androgen receptor-mediated transcription. FEBS Lett 583(12):1880–1886
Rhodes DR, Yu J, Shanker K et al (2004) ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia 6(1):1–6
Kassambara A, Klein B, Moreaux J (2009) MMSET is overexpressed in cancers: link with tumor aggressiveness. Biochem Biophys Res Commun 379(4):840–845
Hudlebusch HR, Santoni-Rugiu E, Simon R et al (2011) The histone methyltransferase and putative oncoprotein MMSET is overexpressed in a large variety of human tumors. Clin Cancer Res 17(9):2919–2933
Okabe H, Satoh S, Kato T et al (2001) Genome-wide analysis of gene expression in human hepatocellular carcinomas using cDNA microarray: identification of genes involved in viral carcinogenesis and tumor progression. Cancer Res 61(5):2129–2137
Lauring J, Abukhdeir AM, Konishi H et al (2008) The multiple myeloma associated MMSET gene contributes to cellular adhesion, clonogenic growth, and tumorigenicity. Blood 111(2):856–864
Garlisi CG, Uss AS, Xiao H et al (2001) A unique mRNA initiated within a middle intron of WHSC1/MMSET encodes a DNA binding protein that suppresses human IL-5 transcription. Am J Respir Cell Mol Biol 24(1):90–98
Kim JY, Kee HJ, Choe NW et al (2008) Multiple-myeloma-related WHSC1/MMSET isoform RE-IIBP is a histone methyltransferase with transcriptional repression activity. Mol Cell Biol 28(6):2023–2034
Keats JJ, Reiman T, Maxwell CA et al (2003) In multiple myeloma, t(4;14)(p16;q32) is an adverse prognostic factor irrespective of FGFR3 expression. Blood 101(4):1520–1529
Marango J, Shimoyama M, Nishio H et al (2008) The MMSET protein is a histone methyltransferase with characteristics of a transcriptional corepressor. Blood 111(6):3145–3154
Sáez B, Martín-Subero JI, Lahortiga I et al (2007) Simultaneous translocations of FGFR3/MMSET and CCND1 into two different IGH alleles in multiple myeloma: lack of concurrent activation of both proto-oncogenes. Cancer Genet Cytogenet 175(1):65–68
Martinez-Garcia E, Popovic R et al (2011) The MMSET histone methyl transferase switches global histone methylation and alters gene expression in t(4;14) multiple myeloma cells. Blood 117(1):211–220
Hudlebusch HR, Theilgaard-Mönch K, Lodahl M, Johnsen HE, Rasmussen T (2005) Identification of ID-1 as a potential target gene of MMSET in multiple myeloma. Br J Haematol 130(5):700–708
Toyokawa G, Cho HS, Masuda K et al (2011) Histone lysine methyltransferase Wolf-Hirschhorn syndrome candidate one is involved in human carcinogenesis through regulation of the Wnt pathway. Neoplasia 13(10):887–898
Pei H, Zhang L, Luo K et al (2011) MMSET regulates histone H4K20 methylation and 53BP1 accumulation at DNA damage sites. Nature 470(7332):124–128
Conflict of Interest
None
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zhou, P., Wu, LL., Wu, KM. et al. Overexpression of MMSET is Correlation with Poor Prognosis in Hepatocellular Carcinoma. Pathol. Oncol. Res. 19, 303–309 (2013). https://doi.org/10.1007/s12253-012-9583-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12253-012-9583-z