Abstract
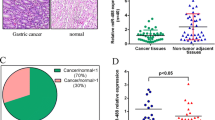
MicroRNAs (miRNAs) are small, non-coding RNAs of endogenous origin. Accumulating studies have shown aberrant miRNA expression plays an important role in many tumor types. miR-192 and -215, which have the same “seed region”, have not been comprehensively investigated using a large number of cases in gastric cancer. The total RNA was extracted from 118 gastric cancer tissues and three gastric cancer cell lines as well as matched non-tumor adjacent tissues (NATs). After polyadenylation and reverse transcription, expression levels of miR-192 and -215 were determined by real-time PCR and calculation using the 2-∆∆CT method for evaluation of the association between miR-192, and -215 expression levels and clinicopathological characteristics. There were no significant differences in miR-192 and -215 expression levels between gastric cancer tissues and non-tumor counterparts (both p > 0.05, paired t-test). Interestingly, miR-192 and -215 were down-regulated in MGC-803 cells, BGC-823 cells and SGC-7901 cells (all p < 0.01, paired t-test). Also, the down-regulation of miR-192 and -215 was demonstrated to be associated with increased tumor sizes (both p = 0.003, Mann–Whitney U test) and advanced Borrmann type tumors (p = 0.015 and p = 0.044, respectively, Kruskal-Wallis H test). Moreover, the expression of miR-192 was significantly lower in the pT4 stage of gastric cancer than in pT1, pT2 and pT3 stages (p = 0.026). Furthermore, there was a strong correlation between miR-192 and -215 in gastric cancer tissues (p < 0.001, Pearson regressions). miR-192 and -215 might be related to the proliferation and invasion of gastric cancer. Potentially, they could become important biomarkers.



Similar content being viewed by others
References
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297
Calin GA, Croce CM (2006) MicroRNA-cancer connection: the beginning of a new tale. Cancer Res 66:7390–7394
Esquela-Kerscher A, Slack FJ (2006) Oncomirs—microRNAs with a role in cancer. Nat Rev Cancer 6:259–269
Hwang HW, Mendell JT (2006) MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer 94:776–780
Cho WC (2007) OncomiRs: the discovery and progress of microRNAs in cancers. Mol Canc 6:60
Calin GA, Sevignani C, Dumitru CD et al (2004) Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A 101:2999–3004
Bandi N, Vassella E (2011) miR-34a and miR-15a/16 are co-regulated in non-small cell lung cancer and control cell cycle progression in a synergistic and Rb-dependent manner. Mol Canc 10:55
Feber A, Xi L, Pennathur A et al (2011) MicroRNA prognostic signature for nodal metastases and survival in esophageal adenocarcinoma. Ann Thorac Surg 91:1523–1530
Guo J, Miao Y, Xiao B et al (2009) Differential expression of microRNA species in human gastric cancer versus non-tumorous tissues. J Gastroenterol Hepatol 24:652–657
Park JK, Henry JC, Jiang J et al (2011) miR-132 and miR-212 are increased in pancreatic cancer and target the retinoblastoma tumor suppressor. Biochem Biophys Res Commun 406:518–523
Chiang Y, Song Y, Wang Z et al (2011) Aberrant expression of miR-203 and its clinical significance in gastric and colorectal cancers. J Gastrointest Surg 15:63–67
Marchini S, Cavalieri D, Fruscio R et al (2011) Association between miR-200c and the survival of patients with stage I epithelial ovarian cancer: a retrospective study of two independent tumour tissue collections. Lancet Oncol 12:273–285
Tsukamoto Y, Nakada C, Noguchi T et al (2010) MicroRNA-375 is downregulated in gastric carcinomas and regulates cell survival by targeting PDK1 and 14-3-3zeta. Cancer Res 70:2339–2349
Chen Y, Song Y, Wang Z, Yue Z, Xu H, Xing C (2010) Altered expression of miR-148a and miR-152 in gastrointestinal cancers and its clinical significance. J Gastrointest Surg 14:1170–1179
Song YX, Yue ZY, Wang ZN et al (2011) MicroRNA-148b is frequently down-regulated in gastric cancer and acts as a tumor suppressor by inhibiting cell proliferation. Mol Canc 10:1
Xiong X, Ren HZ, Li MH, Mei JH, Wen JF, Zheng CL (2011) Down-regulated miRNA-214 induces a cell cycle G1 arrest in gastric cancer cells by up-regulating the PTEN protein. Pathol Oncol Res 17:931–937
Shi R, Chiang VL (2005) Facile means for quantifying microRNA expression by real-time PCR. Biotechniques 39:519–525
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative pcr and the 2 (delta delta c(T)) method. Methods 25:402–408
Du Y, Xu Y, Ding L et al (2009) Down-regulation of miR-141 in gastric cancer and its involvement in cell growth. J Gastroenterol 44:556–561
Shah AA, Leidinger P, Blin N, Meese E (2010) miRNA: small molecules as potential novel biomarkers in cancer. Curr Med Chem 17:4427–4432
Yanaihara N, Caplen N, Bowman E et al (2006) Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Canc Cell 9:189–198
Mathé EA, Nguyen GH, Bowman ED et al (2009) MicroRNA expression in squamous cell carcinoma and adenocarcinoma of the esophagus: associations with survival. Clin Cancer Res 15:6192–6200
Gui J, Tian Y, Wen X et al (2011) Serum microRNA characterization identifies miR-885-5p as a potential marker for detecting liver pathologies. Clin Sci (Lond) 120:183–193
Braun CJ, Zhang X, Savelyeva I et al (2008) p53-responsive micrornas 192 and 215 are capable of inducing cell cycle arrest. Cancer Res 68:10094–10104
Georges SA, Biery MC, Kim SY et al (2008) Coordinated regulation of cell cycle transcripts by p53-Inducible microRNAs, miR-192 and miR-215. Cancer Res 68:10105–10112
Davidson LA, Wang N, Shah MS, Lupton JR, Ivanov I, Chapkin RS (2009) n-3 polyunsaturated fatty acids modulate carcinogen-directed non-coding microRNA signatures in rat colon. Carcinogenesis 30:2077–2084
Baffa R, Fassan M, Volinia S et al (2009) MicroRNA expression profiling of human metastatic cancers identifies cancer gene targets. J Pathol 219:214–221
Jin Z, Selaru FM, Cheng Y et al (2011) MicroRNA-192 and -215 are upregulated in human gastric cancer in vivo and suppress ALCAM expression in vitro. Oncogene 30:1577–1585
Volinia S, Calin GA, Liu CG et al (2006) A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A 103:2257–2261
Lee EJ, Gusev Y, Jiang J et al (2007) Expression profiling identifies microRNA signature in pancreatic cancer. Int J Cancer 120:1046–1054
Quach H, Barreiro LB, Laval G et al (2009) Signatures of purifying and local positive selection in human miRNAs. Am J Hum Genet 84:316–327
Kim DY, Joo JK, Park YK, Ryu SY, Kim YJ, Kim SK (2007) Predictors of long-term survival in node-positive gastric carcinoma patients with curative resection. Langenbeck Arch Surg 392:131–134
Shiraishi N, Inomata M, Osawa N, Yasuda K, Adachi Y, Kitano S (2000) Early and late recurrence after gastrectomy for gastric carcinoma. Cancer 89:255–261
Kim JP, Kim YW, Yang HK, Noh DY (1994) Significant prognostic factors by multivariate analysis of 3926 gastric cancer patients. World J Surg 18:872–878
Pichiorri F, Suh SS, Rocci A et al (2010) Downregulation of p53-inducible microRNAs 192, 194, and 215 impairs the p53/MDM2 autoregulatory loop in multiple myeloma development. Canc Cell 18:367–381
Acknowledgement
This work was supported by National Science Foundation of China (No. 30972879 and No. 81172370), Specialized Research Fund for the Doctoral Program of Higher Education (No. 200801590006) and Natural Science Foundation of Liaoning Province (No. 20092129).
Author information
Authors and Affiliations
Corresponding author
Additional information
Yeunpo Chiang and Xin Zhou contributed equally to this work
Rights and permissions
About this article
Cite this article
Chiang, Y., Zhou, X., Wang, Z. et al. Expression Levels of MicroRNA-192 and -215 in Gastric Carcinoma. Pathol. Oncol. Res. 18, 585–591 (2012). https://doi.org/10.1007/s12253-011-9480-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12253-011-9480-x